Malaria Journal
- Most accessed
- Collections

The unseen battle: interpreting the 2023 World Malaria Report from Burkina Faso's frontlines
Authors: Etienne Bilgo
LLIN evaluation in Uganda project (LLINEUP2): association between housing construction and malaria burden in 32 districts
Authors: Samuel Gonahasa, Martha Nassali, Catherine Maiteki‑Sebuguzi, Jane F. Namuganga, Jimmy Opigo, Isaiah Nabende, Jaffer Okiring, Adrienne Epstein, Katherine Snyman, Joaniter I. Nankabirwa, Moses R. Kamya, Grant Dorsey and Sarah G. Staedke
Space–time clusters and co-occurrence of Plasmodium vivax and Plasmodium falciparum malaria in West Bengal, India
Authors: Meghna Maiti and Utpal Roy
Screening of malaria infections in human blood samples with varying parasite densities and anaemic conditions using AI-Powered mid-infrared spectroscopy
Authors: Issa H. Mshani, Frank M. Jackson, Rehema Y. Mwanga, Prisca A. Kweyamba, Emmanuel P. Mwanga, Mgeni M. Tambwe, Lorenz M. Hofer, Doreen J. Siria, Mario González-Jiménez, Klaas Wynne, Sarah J. Moore, Fredros Okumu, Simon A. Babayan and Francesco Baldini
A five years malaria surveillance data analysis of North Shewa zone, Amhara region, Ethiopia: July 2018 to June 2023
Authors: Tebabere Moltot, Girma Bekele, Zenebe Abebe Gebreegziabher, Tesfansh Lemma, Moges Sisay, Mulualem Silesh, Melkam Mulugeta, Legesse Demissie, Tirusew Nigussie Kebede and Birhan Tsegaw Taye
Most recent articles RSS
View all articles
Plant-based insect repellents: a review of their efficacy, development and testing
Authors: Marta Ferreira Maia and Sarah J Moore
Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria
Authors: Jane Achan, Ambrose O Talisuna, Annette Erhart, Adoke Yeka, James K Tibenderana, Frederick N Baliraine, Philip J Rosenthal and Umberto D'Alessandro
Effectiveness of plant-based repellents against different Anopheles species: a systematic review
Authors: Amin Asadollahi, Mehdi Khoobdel, Alireza Zahraei-Ramazani, Sahar Azarmi and Sayed Hussain Mosawi
Foul wind, spirits and witchcraft: illness conceptions and health-seeking behaviour for malaria in the Gambia
Authors: Sarah O’Neill, Charlotte Gryseels, Susan Dierickx, Julia Mwesigwa, Joseph Okebe, Umberto d’Alessandro and Koen Peeters Grietens
Baseline data of parasite clearance in patients with falciparum malaria treated with an artemisinin derivative: an individual patient data meta-analysis
Most accessed articles RSS
Evaluating the impact of attractive targeted sugar baits in Zambia: a community randomized controlled trial
World Malaria Day - 2024
Cross Journal Collection
Elimination of infectious diseases of poverty as a key contribution to achieving the SDGs
Thematic series Primate malaria
Cross journal collections - closed
Contribution of climate change to the spread of infectious diseases
Human migration, conflict and infectious diseases
Spatial inequality, infectious diseases and disease control
Thematic series Malaria and COVID-19
Thematic series Malaria Elimination Demonstration Project, Mandla, India
Thematic series Durability Monitoring of Long-lasting Insecticidal Nets: Results and Learning from the VectorWorks Project
Thematic series Alternative interventions to facilitate malaria elimination
Thematic series A combined effect: using indoor residual spraying and insecticide-treated nets together for additional impact
Debate series The Malaria Journal debates Edited by Elizabeth Ashley
Thematic series Targeted next generation sequencing for malaria molecular epidemiology in Africa Edited by Jonathan Juliano
Thematic series Time to go for vivax Edited by: Marcus V Lacerda and Hernando A del Portillo
Thematic series Biomarkers of malaria
Cross Journal Collection 14th International Congress of Parasitology
Thematic series ACT now: anti-malarial market complexity one decade after the introduction of artemisinin combination therapy – evidence from sub-Saharan Africa and the Greater Mekong Sub-region
Thematic series Ivermectin to reduce malaria transmission
Thematic series Housing and malaria Edited by: Dr. Lucy Tusting, Dr. Jo Lines and Barbary Willey
Thematic series Re-imagining malaria – a platform for reflections to widen horizons in malaria control Edited by: Dr. Julian Eckl, Dr. Susanna Hausmann Muela
Cross journal collection Every day is Malaria Day
Cross journal collection Reviewer acknowledgements 2013
Thematic series WHO global malaria recommendations 2012 - 2015
Thematic series The ACTwatch project: monitoring anti-malarial markets in seven countries
Thematic series Travellers' malaria Edited by: Prof Patricia Schlagenhauf
Thematic series National malaria control programme (NMCP) Best Practice Sharing Edited by: Prof Robert William Snow
Thematic series Towards malaria elimination
Thematic series The world antimalarial resistance network (WARN)
Latest Tweets
Your browser needs to have JavaScript enabled to view this timeline
World Malaria Day 2024
Collections open for submissions

Cross journal collection

Alternative interventions to facilitate malaria elimination
Malaria Journal In Review - preprints
Malaria Journal , in partnership with Research Square, is offering In Review . Authors choosing this free optional service will be able to:
- Share their work with fellow researchers to read, comment on, and cite even before publication
- Showcase their work to funders and others with a citable DOI while it is still under review
- Track their manuscript - including seeing when reviewers are invited, and when reports are received
See what the Malaria Journal In Review platform looks like
Aims and Scope
Malaria Journal is aimed at the scientific community interested in malaria in its broadest sense. It is the only journal that publishes exclusively articles on malaria and, as such, it aims to bring together knowledge from the different specialties involved in this very broad discipline, from the bench to the bedside and to the field.
Editor-in-Chief - Marcel Hommel, University of Liverpool, UK

- Editorial Board
- Manuscript editing services
- SNAPP Editor link
- Instructions for Editors
- Contact Support for Editors
- Sign up for article alerts and news from this journal
- Follow us on Twitter
Annual Journal Metrics
2022 Citation Impact 3.0 - 2-year Impact Factor 3.2 - 5-year Impact Factor 1.148 - SNIP (Source Normalized Impact per Paper) 1.237 - SJR (SCImago Journal Rank)
2023 Speed 7 days submission to first editorial decision for all manuscripts (Median) 131 days submission to accept (Median)
2023 Usage 4,093,320 downloads 5,053 Altmetric mentions
- More about our metrics
ISSN: 1475-2875
- Submission enquiries: [email protected]
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Review Article
- Published: 08 April 2021
Advances and opportunities in malaria population genomics
- Daniel E. Neafsey ORCID: orcid.org/0000-0002-1665-9323 1 , 2 na1 ,
- Aimee R. Taylor 2 , 3 na1 &
- Bronwyn L. MacInnis 2
Nature Reviews Genetics volume 22 , pages 502–517 ( 2021 ) Cite this article
13k Accesses
53 Citations
19 Altmetric
Metrics details
- Infectious diseases
- Medical genomics
- Parasite genetics
Almost 20 years have passed since the first reference genome assemblies were published for Plasmodium falciparum , the deadliest malaria parasite, and Anopheles gambiae , the most important mosquito vector of malaria in sub-Saharan Africa. Reference genomes now exist for all human malaria parasites and nearly half of the ~40 important vectors around the world. As a foundation for genetic diversity studies, these reference genomes have helped advance our understanding of basic disease biology and drug and insecticide resistance, and have informed vaccine development efforts. Population genomic data are increasingly being used to guide our understanding of malaria epidemiology, for example by assessing connectivity between populations and the efficacy of parasite and vector interventions. The potential value of these applications to malaria control strategies, together with the increasing diversity of genomic data types and contexts in which data are being generated, raise both opportunities and challenges in the field. This Review discusses advances in malaria genomics and explores how population genomic data could be harnessed to further support global disease control efforts.
Similar content being viewed by others

Characterizing the genomic variation and population dynamics of Plasmodium falciparum malaria parasites in and around Lake Victoria, Kenya
Population genomics reveal distinct and diverging populations of An. minimus in Cambodia

Population genetic analysis of Plasmodium knowlesi reveals differential selection and exchange events between Borneo and Peninsular sub-populations
Introduction.
Malaria is a disease caused by parasitic protozoans in the genus Plasmodium , which are transmitted between vertebrate hosts by female mosquitoes in the genus Anopheles . The Plasmodium life cycle involves haploid asexual stages in vertebrate hosts and, in mosquitoes, a diploid stage with sexual recombination between one or more pairs of identical, inbred or outbred gametes 1 . Individual parasite species are usually specialized to infect a narrow range of vertebrate and mosquito hosts. The predominant malaria parasite species that infect humans ( Plasmodium falciparum and Plasmodium vivax ) evolved between 10,000 and 60,000 years ago 2 , 3 , making malaria one of the very oldest human infectious diseases, in addition to one of the most lethal 4 . In 2019, malaria was responsible for more than 400,000 deaths, mostly of young children in sub-Saharan Africa, with an estimated 3.2 billion people around the world susceptible to infection 5 . Although annual malaria mortality has been more than halved in the past 20 years, drug and insecticide resistance, the lack of a highly efficacious vaccine and disruptions such as the COVID-19 pandemic threaten to halt or reverse these gains 5 .
As an ancient disease that inflicts mortality disproportionately on children, malaria has provided a strong evolutionary pressure on human populations to resist infection and disease 4 . Reciprocally, strong evolutionary pressure has been exerted on parasites by the human immune system and antimalarial drugs. As humans have carried malaria parasites around the world, parasites have evolved to enhance their transmissibility by local anopheline mosquito vector species in different regions 6 , which have themselves been under intense selective pressure owing to insecticide use over the past century 7 . This complex co-evolutionary history between humans, Plasmodium parasites and Anopheles mosquitoes is written in the genome of each organism, and genomic tools and data are of key importance for understanding the fundamental genetic underpinning of malaria and developing new strategies to eliminate it. Indeed, every modern malaria intervention, including antiparasitic drugs and insecticides, has been met with an evolutionary response that may be studied through genomic variation. Furthermore, the failure to develop a highly efficacious malaria vaccine is partially attributable to the extreme variability of genes encoding the most immunogenic parasite antigens 8 . Population genomics could therefore be used to help develop effective new interventions, as well as to support the efficient deployment of existing disease control measures through resistance surveillance 9 , tracking the movement of parasites and vectors 10 and measuring changes in transmission intensity using population genomic response indicators 11 .
In this Review, we provide a brief history of the use of population genomics in malaria research and discuss recent advances, focusing on parasites and vectors. We describe innovations for efficient data generation on a large scale and analyses designed to make use of such data for genomic epidemiology. We identify gaps in the availability of reference genome assemblies for anopheline vectors that impede the understanding of key disease-transmitting members of this ancient and diverse mosquito genus. Finally, we make a case for using epidemiological data analysis techniques that consider variation caused by recombination to enable comparisons of diverse genomic data sets and complement and extend conventional analyses of variation that consider differences in allele frequency. This article does not discuss advances in understanding human or vector genomic adaptations to malaria, functional genomics, experimental evolution or parasite or vector transcriptomics, which have been addressed by other recent reviews 12 , 13 , 14 .
Advent of malaria population genomics
The malaria genomics era began with the sequencing and assembly of the West African 3D7 clone of P. falciparum , which was published in 2002 (ref. 15 ) after nearly a decade of collaborative effort between the Sanger Centre (now the Wellcome Sanger Institute), the Institute for Genomic Research (TIGR), the Stanford Genome Technology Center and the Naval Medical Research Center 16 (Fig. 1 ). This 23 Mb reference genome revealed that P. falciparum harbours almost 5,300 genes, including more than 200 genes associated with immune evasion in the var, rifin and stevor gene families, localized to the subtelomeres. More than 60% of the predicted proteins had no sequence similarity with proteins in other organisms and could not be assigned function 15 . Following publication of the P. falciparum genome, multiple groups embarked on characterizing genomic variation in this species, together profiling geographically diverse isolates using Sanger sequencing 17 , 18 , 19 in a template that would later be applied to P. vivax and other parasite species 20 , 21 . These studies identified enough genetic variants to allow targeted and higher-throughput single nucleotide polymorphism (SNP) genotyping of P. falciparum parasites 22 , 23 , 24 to facilitate deeper analyses of selection and demographic history.
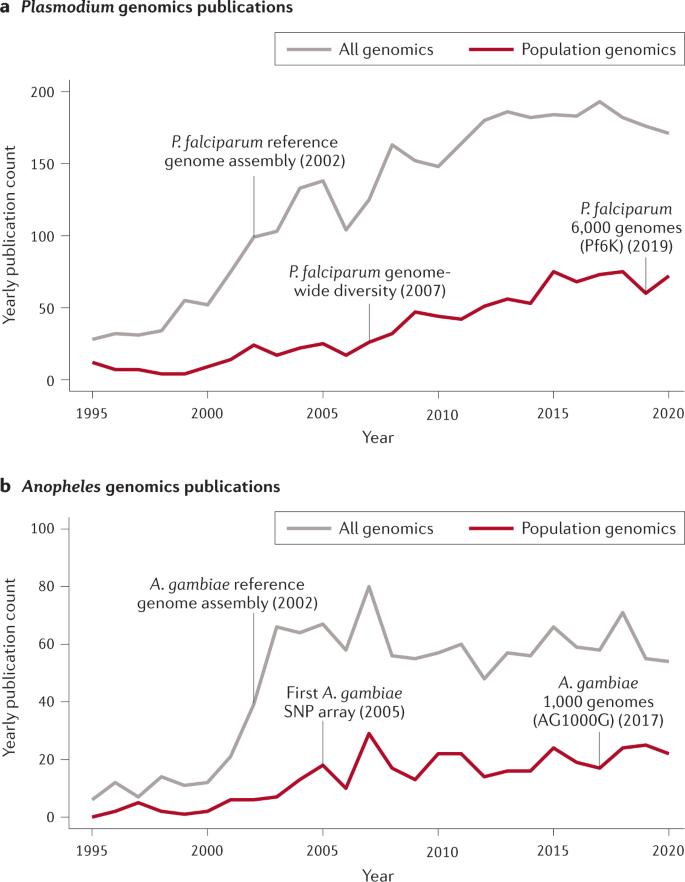
Timelines of Plasmodium and Anopheles genomics publications and highlighted publications. a | Plasmodium genomics publications. An increasing fraction of Plasmodium genomics papers over time also include terms signifying population diversity in the title and/or abstract. Highlighted milestones include the publication of the first Plasmodium falciparum reference genome assembly in 2002 (ref. 15 ), co-publications reporting the characterization of genome-wide diversity in 2007 (refs 17 , 18 , 19 ) and the release of the Pf6K data set in 2019 (ref. 170 ). ‘All genomics’ data represent articles in the NCBI PubMed database published from 1 January 1995 to the present day that include the search terms “Plasmodium” and “genom*” in the title or abstract. ‘Population genomics’ data represent ‘all genomics’ articles that also include the search terms “population”, “epidemio*”, “polymorph*” or “diversity” in the title or abstract. b | Anopheles genomics publications. An increasing fraction of Anopheles genomics papers over time also include terms signifying population diversity. Highlighted milestones include the publication of the first Anopheles gambiae reference genome assembly in 2002 (ref. 25 ), the first genome-wide single nucleotide polymorphism (SNP) genotyping array for A. gambiae in 2005 (ref. 171 ) and the publication of the AG1000G project in 2017 (ref. 29 ). Search queries as above with the replacement of “ Plasmodium ” with “ Anopheles ”.
The first reference genome assembly for a mosquito, Anopheles gambiae , was published in 2002 (ref. 25 ), the same week as the P. falciparum assembly publication and driven by a collaboration between Celera Genomics and investigators at diverse academic institutions. A. gambiae is the most important malaria vector in sub-Saharan Africa and likely the most competent malaria vector in the world owing to its anthropophilic feeding preference, preference for biting indoors and susceptibility to Plasmodium infection 26 . The 278 Mb genome of A. gambiae was found to contain ~14,000 genes, with many related to immunity, olfaction and other traits contributing to malaria vectorial capacity 25 . Although genes encoding the targets of major insecticidal classes had been identified before this sequencing project 27 , 28 , the A. gambiae reference genome assembly provided the first complete catalogue of metabolic genes for any mosquito and would later prove essential in identifying diverse non-target site mechanisms of insecticide resistance in this species 29 , 30 .
Efforts to construct the first A. gambiae reference genome assembly identified extremely high levels of genetic variation. Nearly half a million SNPs were identified among mosquitoes sequenced from pooled DNA, despite the fact that all mosquitoes were derived from a single captive colony 25 . This mosquito colony (known as the PEST colony) was founded as a hybrid population of two cytological forms, Mopti and Savanna, which were later distinguished as the sibling species A. gambiae sensu stricto and Anopheles coluzzii , respectively 31 , which are both part of a species complex that now includes at least eight members 32 . The admixture of these two highly polymorphic sister species in this initial reference assembly project helped to inspire many subsequent studies to distinguish intra-species diversity 18 from genomic divergence between morphologically identical cryptic species 32 , 33 , 34 , 35 , 36 . These studies yielded the first genomic perspective on the tendency of Anopheles mosquito species to rapidly radiate into new taxa, which has been of great interest to biologists studying speciation. As the presence of cryptic species has confounded efforts to measure insecticide resistance phenotypically and genotypically for more than half a century 37 , 38 , 39 , the genomic delineation of morphologically identical species has immense value for disease control.
Reference assemblies for P. falciparum and A. gambiae were just the first steps towards malaria population genomics. Although malaria is referred to as a single disease, the term encompasses infections caused by a range of parasite species that are transmitted by at least 40 species of Anopheles mosquito. Many of these are not closely related sister species within their respective haemosporidian parasite and anopheline mosquito clades (Fig. 2 ). The diverse evolutionary lineages of the anophelines that transmit malaria diverged approximately 100 million years ago 40 as the continents were separating from each other; similarly, the Plasmodium parasite species that infect humans belong to divergent branches of an ancient and polyphyletic genus that comprises species that infect a broad range of primates, rodents, birds, reptiles and ungulates. The deep divergence of parasite and vector lineages makes it difficult to generalize biological knowledge about many aspects of the disease and necessitates the genomic study of its diverse agents. Reference assemblies for many malaria parasites and anopheline mosquito vectors other than P. falciparum and A. gambiae have been described since 2002 (refs 3 , 41 , 42 , 43 , 44 , 45 ) (Fig. 2 ). However, deep explorations of population genomic diversity have not yet been commonly conducted for species other than P. falciparum , P. vivax and A. gambiae/A. coluzzii .
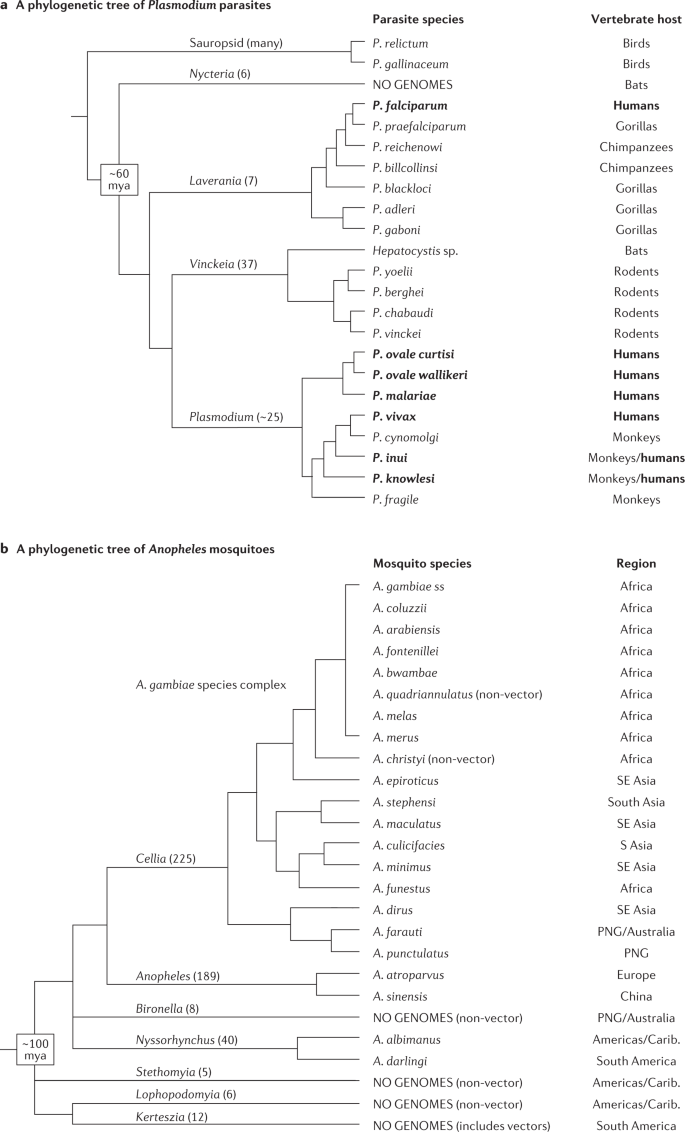
Malaria is a disease caused by various parasite species hailing from diverse subgenera and groups of the ancient and polyphyletic Plasmodium genus (order Haemosporida). Plasmodium parasites are transmitted by different anopheline mosquito vectors, which last shared a common ancestor approximately 100 million years ago (mya) 40 . Numbers in parentheses following subgenus or group labels indicate the approximate number of formally described parasite or mosquito species in each lineage. Reference genome assemblies exist for all species except where indicated. a | A phylogenetic tree of Plasmodium parasites, with topology informed by Galen et al. 172 and Otto et al. 3 . The common ancestor of mammal-infecting members of Plasmodium is estimated to have existed less than 60 mya, as bats are hypothesized to be the ancestral mammalian hosts and this is the estimated date of the Chiroptera radiation 173 . Species that infect humans are in bold. b | A phylogenetic tree of Anopheles mosquitoes with reference to available genomic resources. Topology informed by Foster et al. 174 and Neafsey et al. 42 . Reference genome assemblies are lacking for Kerteszia and other subgenera from the Americas, limiting comparative genomic opportunities to understand vectorial capacity and genomic investigations of insecticide resistance in many vector lineages. PNG, Papua New Guinea; ss , sensu stricto .
In the past decade, advances in malaria genomics have been driven largely by the advent of high-throughput next generation sequencing (NGS) and the use of the reference genomes described above to analyse short reads produced by this approach. The increase in throughput afforded by NGS has expanded malaria genomics applications to include large-scale data sets capturing parasite and vector genetic diversity in natural populations around the world 21 , 29 , 46 , 47 , allowing for new strategies in identifying and tracking drug and insecticide resistance markers 48 , 49 and new approaches to improve our understanding of malaria epidemiology.
Challenges in generating genomic data
Several technical challenges must be addressed before large-scale parasite and vector sequencing can realize its full potential 50 . These challenges include procuring high-quality DNA from parasite and mosquito samples, and contending with the high degree of heterozygosity in vector genomes. Finding solutions to these challenges has been a major focus of innovation in the malaria genomics community and some practical solutions have emerged. We detail some of these challenges and innovations below.
Isolation of parasite DNA
The primary obstacle to producing high-quality parasite genomic data at scale has been obtaining sufficient parasite DNA for sequencing, in part owing to host DNA contamination. In a typical blood sample from a malaria-infected individual there is commonly at least 1,000-fold less parasite DNA than human DNA owing to the disparity in genome size between the host and parasite and a typically low level of parasitaemia. The highly A/T-rich (81%) genome of P. falciparum 15 further exacerbates sequencing coverage issues in clinical samples for this parasite species, as many sequencing library construction methods are biased against incorporation of highly A/T-rich DNA fragments 51 . It is therefore often a costly and inefficient exercise to simply sequence total DNA from infected blood samples to obtain parasite genomic data.
In vitro cultivation of Plasmodium species, including P. falciparum , can facilitate access to pure parasite DNA 52 . However, in vitro parasite culture is laborious and risks introducing artefacts into the genetic diversity profile from the original natural infection as mutations can be selected for by growth under in vitro culture conditions 53 . Furthermore, other parasites that infect humans, such as P. vivax , Plasmodium malariae and Plasmodium ovale have proved difficult to adapt to in vitro culture 54 .
The development of methods for obtaining enriched parasite DNA from infected blood samples without the need for in vitro culture has been driven by the increased capacity of NGS for profiling population samples. These methods include column-based host leukocyte depletion prior to DNA extraction 55 , hybrid selection of extracted DNA using oligos complementary to the parasite genome 56 , 57 and selective whole-genome amplification (SWGA) of extracted DNA using priming oligos specific to the parasite genome 58 , 59 , 60 . Practically, hybrid selection and SWGA enable the sequencing of blood spots from finger prick samples and avoid the need for the high-volume venous blood collections required for leukocyte depletion, better facilitating sampling for population genomic studies. SWGA has now been widely implemented, although it does not produce spatially uniform sequencing coverage across parasite genomes or usable depths of sequencing coverage from samples with degraded DNA 9 , 61 , 62 .
Obtaining high-grade vector DNA
Many key vector species lack reference genome assemblies, which is a barrier to the application of population genomic approaches to malaria in many regions of the world. High-quality reference assemblies are needed for many secondary (or locally primary) vector species in Africa, including Anopheles moucheti and Anopheles nili 63 . In Latin America, Southeast Asia and the Southwest Pacific, many primary and secondary malaria vectors lack high-quality reference genome assemblies and indeed may not be recognized taxonomically 64 , 65 , 66 , 67 , 68 . The key to good mosquito assemblies, whether sequencing data are based on short or long reads, is high molecular weight DNA. The most effective method for preserving mosquitoes in order to keep high molecular weight DNA intact for sequencing involves flash freezing them in liquid nitrogen and shipping them on dry ice if sequencing facilities are not locally available. However, liquid nitrogen is not available in many settings and international dry ice shipments are expensive and logistically complex. Until sequencing capacity becomes further decentralized, simple and cost-effective means to preserve high molecular weight DNA in mosquitoes are needed to facilitate sample collection in regions without access to these specialized resources.
Addressing vector heterozygosity
As mentioned above, producing reference genomes for Anopheles mosquitoes has been hampered by the high degree of heterozygosity in Anopheles genomes. A recent effort to characterize diversity in 765 wild A. gambiae sensu stricto and A. coluzzii mosquitoes caught at numerous collection sites in Africa identified more than 52 million SNPs in the 141 Mb region of the genome most accessible to accurate variant calling — a rate of one variant allele every 2.2 nucleotides 29 . This degree of heterozygosity is striking among eukaryotes and presents a challenge for genome assembly algorithms that are geared towards the much lower levels of heterozygosity typical of humans and other vertebrates. The separate assembly of maternal and paternal haplotypes into distinct haplotigs can occur when divergence between similar assembly fragments is mistakenly inferred to represent paralogy (duplication) rather than heterozygosity and if not recognized can inflate the size of a haploid reference assembly. However, haplotype-aware graph-based assembly algorithms have improved tolerance of heterozygosity and are being applied to new anopheline assembly projects 69 , 70 . Further, the DNA input requirements for long-read sequencing have become smaller in recent years, enabling long-read sequencing libraries to be made from a single mosquito instead of a pool of mosquitoes, reducing genetic variation in the sequencing template 71 .
Using malaria population genomic data
The creation of reference assemblies and polymorphism data sets for malaria parasites and vectors has improved our understanding of diverse aspects of malaria biology over the past 15 years, and genomic data from natural populations could inform control and elimination strategies. We feature some uses of such data sets below.
Characterizing genomic diversity
Genomic variation has been characterized in various ways in parasites and vectors. ‘Pre-genomic’ approaches developed before reference genome assemblies were available, such as microsatellites 72 and allele-specific PCR 73 with agarose gel profiling of PCR amplicon size polymorphisms 74 , continue to find use owing to their simplicity and low cost. Post-genomic approaches include small SNP genotyping panels 75 , 76 , 77 , 78 , large SNP genotyping arrays 23 , 79 , targeted sequencing of amplicons 80 , 81 , 82 , 83 , 84 or molecular inversion probes (MIPs) 85 and shotgun whole-genome sequencing (WGS). These approaches 46 , 86 , 87 , 88 are summarized in Box 1 .
Large-scale resequencing studies identifying SNPs across the genomes of P. falciparum and P. vivax in blood samples from around the world have shown clear population structure within and between countries and continents 19 , 21 , 46 , 47 , 88 . The extent and nature of parasite genomic diversity reflects prehistoric transmission levels, human and parasite movement during the colonial era and population bottlenecks caused by control efforts in the last century 21 , 46 , 47 , 50 , 88 , 89 . Similar efforts characterizing the genetic diversity of malaria vectors across Africa — primarily A. gambiae and A. coluzzii — revealed incredible population diversity and structure across the continent 29 . Such resequencing studies provide a framework for the development of geographically targeted genotyping panels where WGS is not feasible. Key applications of these large-scale catalogues of diversity are described below.

Box 1 Generating population genomic data in Plasmodium and Anopheles
In addition to whole-genome sequencing, multiple other technologies are used for targeted sequencing or genotyping of parasites and vectors.
msp/glurp genotyping
Nested PCR assays have been used to detect polymorphisms in the genes msp1 and msp2 , which encode highly polymorphic Plasmodium falciparum merozoite surface proteins 1 and 2 and the gene glurp encoding glutamate-rich protein. Common applications include estimation of complexity of infection in population genetic studies and to distinguish recrudescent infections from new infections in therapeutic efficacy studies 73 , 74 . Although these methods are straightforward and inexpensive, they provide lower resolution than sequencing approaches.
Microsatellite genotyping
A staple genotyping approach for diverse organisms, including Plasmodium and Anopheles , particularly in the pre-genomic era 143 , 176 . Amplification and sequencing or gel-based characterization of tandem repeats exhibiting length polymorphism gives a readout of allele length, with a high per-locus count of genetic variants. Imprecision in allele length estimation can complicate interpretation and limits the scalability of microsatellite marker panels. Microsatellites were used to identify the intercontinental spread of P. falciparum antimalarial drug resistance alleles from Asia to Africa 72 and more recently, across the eastern Greater Mekong Subregion 177 , 178 .
SNP genotyping
Diverse technologies exist for genotyping single nucleotide polymorphism (SNP) loci previously discovered through whole-genome sequencing. The malaria genomics field has employed approaches including Sequenom/Agena, Taqman and high-resolution melting curves for genotyping single biallelic SNPs in large numbers of parasite or vector samples 179 . SNP genotyping is inexpensive at scale but requires specialized instrumentation. SNP genotyping microarrays first gave insight into the chromosomal architecture of speciation between Anopheles gambiae and Anopheles coluzzii 171 .
PCR amplicon sequencing
Next-generation sequencing (NGS) of short PCR amplicons (100–300 bp) has been used in the malaria genomics field for monoplexed and multiplexed PCRs to genotype single variants and characterize ‘microhaplotypes’ of multiple linked variants 82 , 85 . If amplicons are targeted to short windows exhibiting a high density of SNPs, allelic richness at the haplotype level can be high, even if component SNPs are merely biallelic. Amplicon sequencing of highly haplotypically diverse antigens has provided insight into complex infections 82 and vaccine efficacy 122 , and has been applied for surveillance of known and novel alleles at drug resistance loci 180 , 181 .
Capture/hybridization-based sequencing
Hybrid capture of the whole parasite genome has been used to perform selective sequencing of parasite DNA in clinical samples dominated by host DNA 56 , 57 and may be employed for targeted sequencing of informative marker panels within parasite and vector genomes. Molecular inversion probes (MIPs) have been applied to capture, amplify and sequence thousands of short targets (100–150 bp) in parasite genomes. MIP capture and sequencing are well-suited for targeted sequencing of more genomic regions than multiplexed amplicon sequencing, but are less sensitive. MIPs recently provided a detailed profile of drug resistance allele prevalence and P. falciparum population connectivity in the Democratic Republic of the Congo 182 .
Identification and monitoring of parasite drug resistance loci
Malaria parasites have evolved resistance to all widely deployed antimalarial drugs 90 . In some cases, resistance has spread globally, whereas for other compounds it remains rare and patchily distributed. Retrospective genomic resistance surveillance can inform understanding of how drug resistance emerges, long after the opportunity to directly measure patient phenotypes has passed and with greater resolution than individual locus-based surveys 91 . Prospective genomic surveillance can identify signatures of emerging drug resistance in the form of selective sweeps even before specific resistance loci are identified 92 . Parasite resistance to chloroquine helped to undermine the WHO Global Malaria Elimination Campaign of 1955–1969 (ref. 93 ). In 2000, the gene primarily responsible for mediating chloroquine resistance, pfcrt — which encodes an ATP-binding cassette transporter — was identified in parasites 94 . Subsequent resistance surveillance efforts revealed that resistance to chloroquine spread around the globe after arising de novo in Southeast Asia and South America 95 . Resistance to sulfadoxine/pyrimethamine 72 — the successor to chloroquine as a first-line therapy — also emerged first in Southeast Asia and South America and was transported to Africa before the genes associated with resistance (dihydrofolate reductase, pdhfr , and dihydropteroate synthase, pfdhps ) could be discovered 96 .
Resistance has recently arisen to artemisinin 97 , 98 , the current first-line therapy (in combination with various partner drugs) for P. falciparum malaria in much of the world. In vitro resistance selection and WGS of a parasite line showing reduced in vitro susceptibility to artemisinin led to the identification of molecular markers for artemisinin resistance 99 . Mutations in the propeller domain of the Kelch domain-containing protein PfK13 (encoded by a gene on chromosome 13, pfkelch13 ) and especially a C580Y mutation in that domain have been corroborated as agents of reduced susceptibility to artemisinin. Genomic evidence for this association has arisen from allelic replacement studies 100 , genome-wide association studies 48 , 101 , linkage group selection experiments 102 and selection scans 92 , 103 , 104 . Global genomic surveillance is ongoing for pfkelch13 mutations and for epistatic or fitness-compensating mutations at loci elsewhere in the P. falciparum genome, including resistance mutations for partner drugs used in combination therapy 105 . Although pfkelch13 mutations have been confirmed to occur de novo in geographical locations outside their first origin in the Greater Mekong Subregion (GMS) 61 , 62 , 106 , 107 , surveillance has shown that they are still rare in sub-Saharan Africa, South America and New Guinea, raising the hope that retrospective 91 and current genomic surveillance efforts 85 , 88 , 108 , 109 could limit the global spread of resistance to artemisinin-based combination therapies by guiding locally effective treatment policies informing resistance management efforts. For example, given that population genomics has demonstrated that the emergence of the pfkelch13 C580Y allele both inside and outside the GMS has been driven by dozens of recurrent de novo mutations 61 , 62 , 91 , 104 , 106 , a strategy of resistance containment in the GMS without balanced resistance surveillance efforts elsewhere may not be viable over the long term 110 , 111 , 112 .
Identification and monitoring of mosquito insecticide resistance loci
Population genomics has the potential to enable evidence-based management of insecticide resistance in mosquito vectors 113 . The majority of progress in reducing malaria mortality during the past 20 years is attributable to vector control 114 , and anopheline mosquitoes have collectively evolved resistance to all four of the chemical classes of insecticide currently used for control at the adult stage, namely pyrethroids, organochlorines, carbamates and organophosphates 113 . Mutations in genes encoding the binding targets of insecticides were discovered early on using PCR at loci known to confer resistance in agricultural pests: knockdown of vgsc , a gene encoding a voltage-gated sodium channel, conferred resistance to pyrethroids and organochlorine insecticides such as DDT 27 , whereas knockdown of the acetylcholinesterase gene ace1 conferred resistance to carbamates and organophosphate compounds 28 , 115 .
Whole-genome-based approaches have revealed a fascinating array of resistance mechanisms outside of insecticide binding targets, including metabolic resistance encoded by mutations in or upregulation of cytochrome P450 genes and glutathione S -transferase (GST) genes 116 ; barrier resistance through modification of the thickness or properties of the insecure cuticle 117 ; behavioural resistance — for example, avoidance of insecticide-treated nets or walls — and other more specific mechanisms, such as the overexpression of a sensory appendage protein (SAP2) that binds with high affinity to pyrethroids 118 . Population genomic data have revealed how diverse adaptations have been achieved through SNPs and gene duplications and other structural variations 30 and how such mutations have introgressed among closely related species in different locales 29 . For example, copy number variants occur in all five of the major clusters of A. gambiae gene families associated with metabolic resistance, with high population frequencies and haplotypic structure suggesting that they have been spread by positive selection 30 . This observation refutes the notion that resistance is diagnosable by simple genotyping of SNPs in target genes and suggests that WGS and improved bioinformatic methods for copy number variant detection will be invaluable in tracking insecticide resistance 30 .
Aiding vaccine development
Population genomic approaches have shown that parasite antigenic diversity plays a role in reducing vaccine efficacy 119 . Development efforts began for many candidate P. falciparum vaccines during the pre-genomic era, when the immunogenicity of parasite targets had been measured but not their diversity. Although P. falciparum exhibits moderate genomic diversity overall, many of the genes encoding the antigens that have been the focus of most malaria vaccine development efforts, such as those for circumsporozoite protein (CS), apical membrane antigen 1 (AMA1) and thrombospondin-related adhesion protein (TRAP), exhibit extremely high polymorphism (Fig. 3 ). Most monovalent peptide vaccines based on these antigens have shown limited-to-moderate efficacy in clinical trials to date, in part because they have targeted a single parasite allele and have elicited imperfect cross-protective immunity to other parasite alleles 120 . In a phase II trial of a monovalent peptide vaccine targeting the AMA1 protein of P. falciparum , overall efficacy was observed to be 20%, whereas efficacy against infections by parasites with an allele matching the vaccine allele was 64% 121 . Similarly, in a phase III trial of RTS,S/AS01 — the first and only malaria vaccine to be licensed — vaccine efficacy against infections by parasites with an allele matching the vaccine construct at the P. falciparum CS protein was 50%, compared with 33% for infections by parasites with a non-matching allele 122 . It is presumed that this high degree of diversity in these genes has been maintained through parasite population bottlenecks or has been subsequently restored through immune-mediated balancing selection, driven by highly allele-specific immune protection 123 .
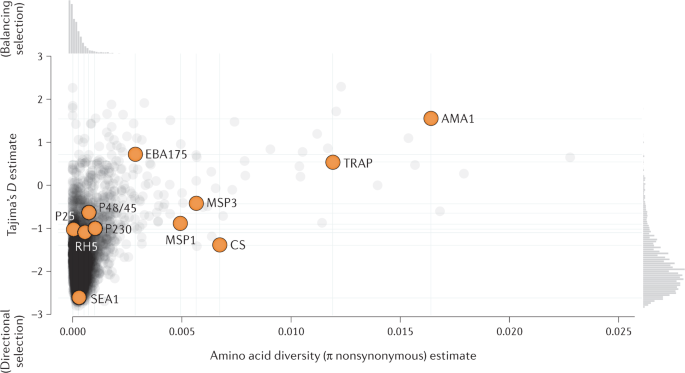
Many monovalent protein subunit vaccine development efforts began before the extensive diversity of many blood-stage and liver-stage antigens was appreciated. Each dot represents a P. falciparum protein; semi-transparent grey dots indicate proteins not targeted by vaccines, opaque orange dots indicate protein targets of vaccines registered for clinical trials at ClinicalTrials.gov since 2000 (ref. 175 ). The horizontal axis indicates estimates of mean amino acid nonsynonymous pairwise diversity, π. The vertical axis depicts estimates of Tajima’s D , a neutrality statistic based on the variant site frequency spectrum, where negative values represent an excess of low-frequency variants and positive values represent an excess of high-frequency variants, which can indicate balancing selection. Estimates were computed using a collection of sequenced P. falciparum parasites from Malawi in the Pf3k resource . Compared with non-vaccine candidates, vaccine candidates generally exhibit a higher level of diversity (Wilcoxon rank sum test, W = 8322.5, P = 0.0001) and show evidence of balancing selection (Wilcoxon rank sum test, W = 8967, P = 0.0002). The newer, post-genomic blood-stage vaccine target RH5 exhibits considerably less diversity, as do several targets of transmission-blocking vaccines (P230, P25 and P48/45).
Population genomics will play an important role in future vaccine development efforts by mapping differences in vaccine target polymorphism profiles across populations and identifying immunogenic protein targets that are not highly polymorphic in any geographical region. Interestingly, several targets of so-called transmission-blocking vaccines (P230, P25 and P48/45), which do not aim to protect the recipient but instead to reduce the likelihood of onward transmission 124 , show low levels of diversity relative to most targets of infection-inhibiting vaccines in parasite populations (Fig. 3 ), perhaps owing to reduced selection by natural immunity. In addition, the P. falciparum reticulocyte binding protein homologue 5 (RH5) protein plays an important role in red blood cell invasion, and population genomic data show that it exhibits low diversity in parasite populations 125 , indicating that a vaccine targeting RH5 may avoid issues with allele-specific protection. Antibodies raised to RH5 have been found to have strong inhibitory activity against P. falciparum 126 . Similarly, a monoclonal antibody that binds to an epitope in the P. falciparum CS protein that lies near the junction between the N-terminal region of the protein and a repetitive region has been shown to protect against infection 127 . This epitope exhibits very low diversity in parasite populations, which has fuelled efforts to evaluate protection conferred by antibodies targeting this region 128 .
Understanding infection and transmission
Understanding transmission intensity and dynamics has historically relied on conventional measures such as case incidence and prevalence, and information from patient travel histories. While these data are crucial, genomic data from parasites in natural populations have the potential to improve understanding of malaria transmission and guide strategies to control it 10 , 11 , and further research is needed to understand the nature, generalizability and utility of genomic signatures as indicators of transmission intensity.
Although malaria parasites are haploid single-celled organisms — apart from a brief diploid stage in the mosquito — populations of malaria parasites may exhibit considerable genomic heterogeneity within single human and mosquito hosts and this heterogeneity can provide insight into the nature and intensity of disease transmission. Efforts to study parasite genomic variation within mosquitoes at drug resistance markers and other loci of interest are in their early stages 129 , 130 ; however, the diversity of parasites within human hosts has been well characterized using microsatellite and msp/glurp genotyping in the pre-genomic era and, in recent years, using NGS of PCR amplicons and single-cell WGS. Deep sequencing of PCR amplicons using NGS allows sensitive characterization of parasite genetic diversity within the host; this is especially effective when the amplicons target regions with high haplotypic diversity, as it is unlikely that distinct parasites within a host will harbour the same sequence at the target region 80 , 81 , 82 , 83 .
An investigation of declining local transmission in the Thailand–Myanmar border region found that the strongest genetic signal associated with transmission decline was loss of parasite genetic diversity within hosts, as measured by a decline in the proportion of complex infections 131 — suggesting that this measure could be a proxy for local transmission intensity 132 . Complex infections are infections that contain parasites with more than one distinct genomic sequence, where the number of distinct genomic sequences is the complexity of infection (COI). Complex infections may result from separate mosquito bites in a process called superinfection and/or from a single mosquito bite in a process called co-transmission (Fig. 4 ). Ascertaining levels of transmission intensity from COI estimates requires assumptions about how new complex infections are founded. Genetically distinct parasites may be related (share some recent common ancestry) or unrelated. As superinfection is expected to be more frequent in high-transmission settings where there is typically a large diversity of parasites, parasites transmitted in different bites are likely to be unrelated. Meanwhile, genetically distinct parasites that are co-transmitted are more likely to be related than unrelated because there are many ways of sampling genetically distinct human-infective sporozoites following recombination between one or more gamete pairs in a single mosquito, yet unrelated sporozoites can be sampled only if two or more unrelated gamete pairs have selfed.
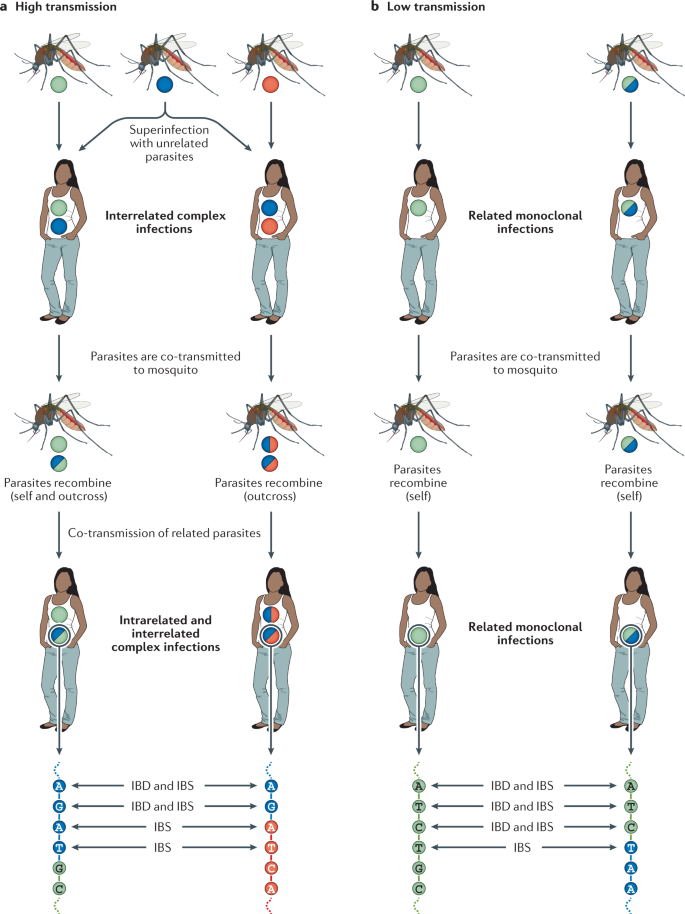
Complex infections result from multiple mosquito bites (superinfection) and/or from a single mosquito bite (co-transmission). Recombination between genetically distinct malaria parasites can occur after a mosquito feeds on a host with a complex infection; otherwise, reproduction occurs by selfing. Genomic data may be used to observe loci that share the same allele (identical by state (IBS)) and to infer loci that are descended from a recent common ancestor (identical by descent (IBD)). a | High transmission, showing three mosquitoes, each infected with parasites from a single lineage, founding interrelated complex infections in two individuals through superinfection. Gametocytes that develop within these complex infections are imbibed by mosquitoes in which they subsequently recombine. Two illustrative scenarios are shown: one in which both selfing and outcrossing occurs, another in which only outcrossing occurs. These mosquitoes founded intra- and interrelated complex infections in two subsequent individuals, this time through co-transmission. b | Low transmission, showing the clonal propagation of two distinct but related parasite genomic sequences following selfing in the mosquito. Each circle represents one or more parasites that share the same genomic sequence. Different circles represent distinct genomic sequences; different colours represent different genetic lineages.
Both co-transmission and superinfection events can be experienced by a single host, thereby giving rise to complex infections with both genetically related and unrelated parasites. Until recently, co-transmission was considered unlikely to account for most complex infections. This is because, with each infectious bite, various bottlenecks reduce the number of surviving parasites and thus the number of distinct genomic sequences among the surviving parasites. For example, mosquitoes typically inject only 10–100 sporozoite-stage parasites per bite 133 . As few as 20% of these sporozoites may exit the dermal inoculation site 134 and only a small fraction of these are likely to avoid clearance by the spleen and immune responses to successfully infect the liver and establish a blood-stage infection 135 . However, recent studies using single-cell WGS of parasite-infected host red blood cells suggest that serial co-transmission of parasites may be common 136 , 137 , 138 . Profiling of parasite genomes from hundreds of single parasite-infected cells identified levels of recent common ancestry and recombination among parasites within hosts that are considered too high to be explained by superinfection alone, indicating multiple rounds of co-transmission of related parasite lineages between hosts 136 , 137 , 138 and raising questions about whether new infections are typically founded by more sporozoites than previously assumed. Single-cell sequencing studies of parasite genomic diversity at the sporozoite stage within mosquito salivary glands are needed to investigate whether malaria infections are founded by a larger inoculum of parasites than previously thought.
Enabling genomics for malaria control
Several applications for malaria genomic data involve spatially differentiating parasite populations based on their genetic profile 139 . The distinct genomic signatures of parasites allow investigation of the connections between parasite populations to identify previously unappreciated sources and sinks of infections and to assess whether cases in low-transmission areas are caused by local transmission or importation — scenarios that demand distinct intervention strategies and the differentiation of which has implications for achieving or maintaining official malaria elimination status. Meaningful implementation of malaria genomics approaches for public health will require careful consideration of the current operational challenges 140 , including sustainable data generation and analysis capacity, harmonization of data and analysis approaches among localities, reagent supply chains, timelines and cost, and the practical value of the information itself.
Data generation
The capacity to generate and analyse local or regional data will be key to the sustainability of applied malaria genomic epidemiology and to ensure efforts are responsive to local context and needs. Efforts to establish genomic surveillance within malaria-endemic countries are increasing rapidly as data generation technologies advance and sequencing costs have fallen 141 . There is an emerging consensus that decentralized data generation capacity within malaria-affected countries or in regional hub labs will enable customization of approaches to local questions and capacities, faster turnaround and closer integration with existing malaria control efforts. Several successful demonstrations of the viability of this model exist at individual institutions, and momentum is building through continent-wide endeavours such as the Africa CDC’s Pathogen Genomic Intelligence initiative and grassroots collaborative organizations such as the Plasmodium Diversity Network Africa (PDNA) 50 , 88 .
Although sequencing costs have fallen markedly over the past two decades, large-scale WGS of parasite and vector samples for surveillance purposes is still impractical in many settings. The additional cost and technical challenge associated with the storage and processing of WGS data relative to targeted genotyping panels are likely to result in a reduced return on investment in a decentralized data generation system 50 , 109 . WGS is essential for identifying new genetic variants relevant for surveillance and validating genomic methodological advancements; however, the research community has been actively evaluating the potential role of targeted sequencing and genotyping approaches to recover the most informative genomic regions for malaria genomic epidemiology 9 , 140 , 142 . On the basis of this work, we suggest that for some applications, deep (1,000×) sequencing coverage of small genomic windows — comprising the most informative 20 kb of malaria parasite genomes, such as highly diverse regions and known markers of interest — could be equally or more operationally useful than 100× sequencing coverage of the whole 23–27 Mb parasite genome and require 100-fold lower net data production per sample. Given that the malaria vector genome is tenfold larger than the parasite genome, there is even greater incentive for targeted sequencing for mosquitoes. Evaluation of which approaches are best suited to in-country use must consider the questions being addressed, the cost and maintenance requirements of instruments and reagents, interpretation of the resulting data and the direct value of the resulting data for informing actionable decisions regarding disease control.
Data analysis
The proliferation of sequencing and genotyping approaches and the decentralization of data generation serve the needs of diverse users and applications but complicate the issue of data interoperability between different research and public health efforts. Descriptive statistics that summarize allele frequency variation within populations or across populations vary with data type, as different categories of polymorphism, for example, SNPs and microsatellites, have mutation rates that vary by 1,000-fold 143 . As such, these widely used statistics, which include π — a measure of intra-population diversity 144 that is often used as a correlate of population size 145 — and the fixation index ( F ST ) — a measure of inter-population differentiation 146 that is sometimes used as a correlate of gene flow 147 — are inappropriate for studies and meta-analyses that feature different input data types. Some convergence in methodology between data generators is likely, although as technology for generating genomic data for malaria epidemiology advances this could lead to further data interoperability issues over time, potentially impeding a longitudinal perspective on malaria even within geographical regions.
Relatedness-based analysis to bridge genomic data types
Relatedness is a recombination-based measure of recent shared ancestry between two individuals 148 . It is an example of a measure that can be compared across different categories of genotypic polymorphism. Although it is not possible to estimate relatedness between samples of different data types, it might be possible to leverage the transitive nature of relatedness and/or the nested nature of some data types, such as SNP barcodes within WGS data, to compare some samples across disparate data sets — an opportunity for future research.
Relatedness is a useful measure for malaria genomic epidemiology because it is recombination based. Unlike π and F ST , which reflect variability in allele frequencies, relatedness is broken down by outcrossing recombination, ranging from one for identical clones to zero for completely unrelated organisms. For a pair of haploid malaria parasites, relatedness is defined as the probability that, at any given locus on the genome, the two alleles are identical by descent (IBD) 149 . For diploid mosquito vectors, there are numerous relatedness measures depending on the pattern of IBD among the four alleles at any locus 150 . Alleles are IBD if descended from a recent common ancestor in some ancestral population; identity by descent can also be defined in terms of chromosomal segments, descended intact, unbroken by recombination, from a recent common ancestor 151 . IBD segments can be used to detect regions of the genome under selection by drugs or other unknown agents 88 , 89 , 91 , 152 . Based on their length, they can also be used for telling closely related and outbred pairs apart from more distantly related but inbred pairs with the same relatedness, although there is a wide distribution of lengths among recent generations 153 . However, IBD segment inference does not solve the issue of achieving commensurate inferences between data types as segment inference specifically requires WGS data 149 , whereas relatedness may be inferred under a statistical model using more modest data 152 , 154 (Fig. 5 ).
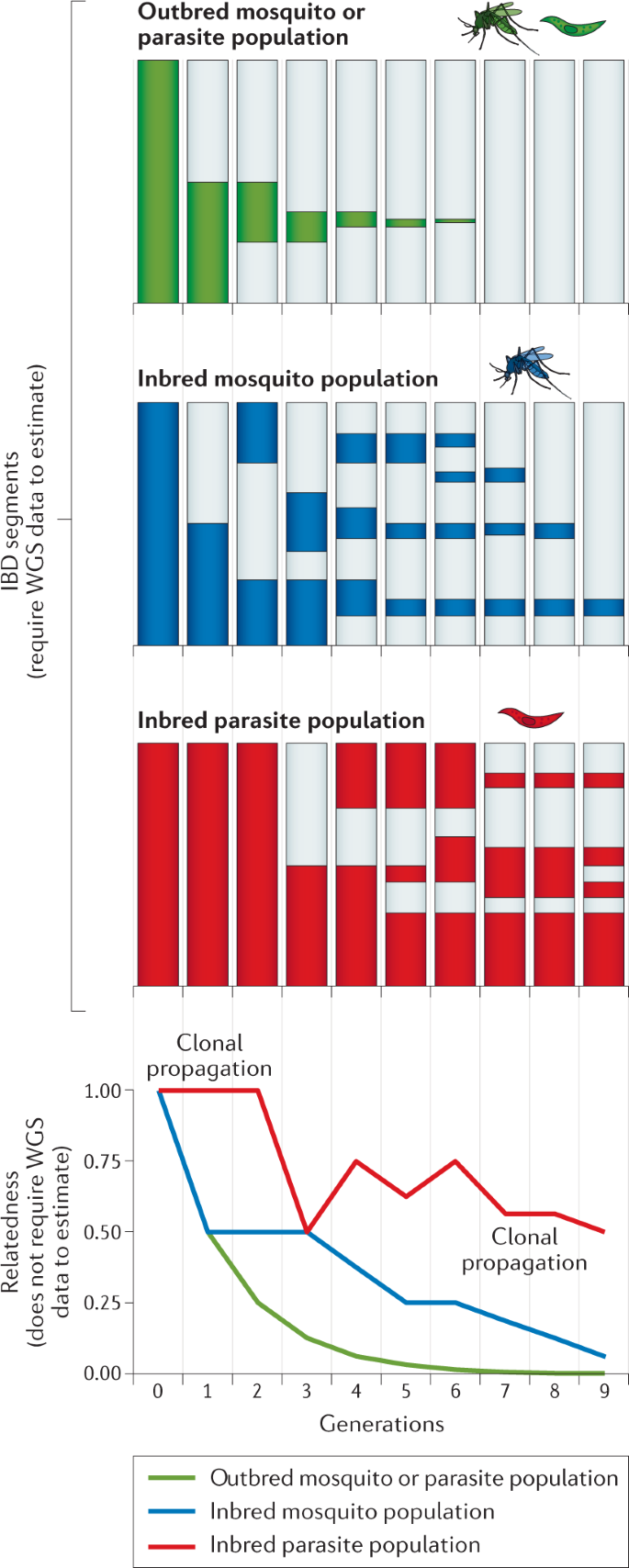
An illustrative example of how recombination over successive generations breaks down chromosomal segments of identity by descent (identical by descent (IBD) segments) and decreases relatedness (probability of IBD). IBD segments, used in various applications including selection detection, require whole-genome sequencing (WGS) data to estimate, whereas relatedness values, used in applications including population connectivity characterization, do not require WGS data to estimate. The top three panels show the breakdown of IBD segments over generations when a mosquito or parasite from one population migrates into mosquitoes or parasites from another population. Vertical bars represent chromosomal genomic sequence of an individual (mosquito or parasite) sampled in the receiving population. IBD between the original migrant and the sampled individual is represented by coloured fill. Light grey represents sequence that is not IBD. Generation zero represents a comparison between the original migrant and itself. Thereafter, we show IBD between the migrant and its closest relative. Segments of IBD break down over successive generations; breakdown depends on the opportunity for outcrossing, which is limited in inbred populations, especially parasite populations, because parasites intermittently self. For each of the situations described in the top three panels, the bottom panel shows the corresponding decrease in relatedness.
Relatedness estimates based on different data types are comparable to each other and can facilitate direct comparisons of distinct studies, provided that measurements of uncertainty are used to account for variations in informativeness across different data types. Observations of high relatedness should be interpreted differently for populations with different rates of inbreeding and selfing, however, as high relatedness is intuitively less likely to imply recent shared ancestry in inbred populations. High relatedness thresholds have been used to make informative contrasts between relatedness estimates within studies, and early efforts towards threshold-free approaches may help to make these summaries more commensurate across studies 155 . It is important to note that, between two individuals, the fraction of markers that are identical by state (IBS) — those that share the same sequence — is a correlate of relatedness; however, IBS-based measures of relatedness do not account for differences in allele frequencies and thus are not commensurate across data types 149 .
Applications of relatedness for malaria
Estimation and interpretation of relatedness between pairs of samples can be applied to multiple epidemiological use cases. Where the outcrossing recombination rate is sufficiently high, IBD breaks down more rapidly than allele frequency variation accumulates and thus measures based on IBD, including relatedness, can be used to characterize changes in parasite or vector population demography more rapidly and at a higher spatial resolution than measures based on allele frequency variation such as π or F ST (Fig. 6 ). In genomic epidemiology, the fast mutation rates of RNA viruses and other measurably evolving pathogens, which generally do not sexually recombine, are used in phylogenetic inference to study pathogen transmission 156 . Plasmodium parasites exhibit SNP mutation rates typical of eukaryotes, on the order of 10 −9 (ref. 157 ) to 10 −10 (ref. 158 ) substitutions per site per generation. It is possible to infer malaria transmission networks over short timescales using sensitive sequencing and analysis techniques to capture rare de novo variants 159 . However, this is not as practical for malaria as it is for viruses and bacteria 156 because of the slow mutation rate of Plasmodium parasites compared with these pathogens, because Plasmodium parasites sexually recombine — complicating phylogenetic inference — and because of insufficiently dense sampling of transmission chains in higher burden settings. As for allele frequency variations above, mutations do not occur quickly enough to allow differentiation of malaria parasite movements between populations separated by distance scales of tens of kilometres or timescales of months to years, whereas detection of relatedness between samples can allow population connectivity measurement at these spatiotemporal scales 10 , 160 to delineate unconnected population units and rapidly inform disease elimination interventions.
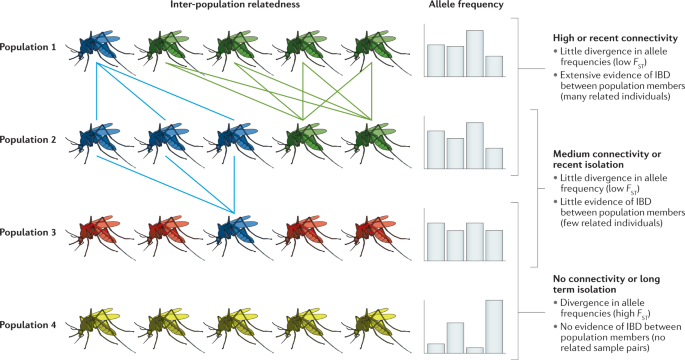
An illustrative example of how relatedness, a measure of the probability of identitical by descent (IBD), may reveal connectivity between populations of mosquitoes on a timescale more relevant for epidemiological applications than allele frequency variation, using genetic differentiation, the fixation index ( F ST ), as an example measure of allele frequency variation. Rows represent distinct mosquito populations. Mosquitoes represent individuals sampled, coloured by their dominant lineage, and edge connections indicate high relatedness and therefore high probability of IBD between pairs of mosquitoes across populations. Populations with a higher degree of connectivity are liable to share more highly related edges. Variability in relatedness between populations may occur with or without appreciable inter-population variation in the allele frequency of genotyped markers. Allele frequencies are indicated by histograms, with vertical axes depicting allele frequencies and horizontal axes depicting markers. F ST describes variation in allele frequency between populations, with poorly connected populations expected to exhibit more vicariant allele frequencies. Genetic drift, migration and/or selection generally take much more time to shift minor allele frequencies among isolated populations relative to recombination, which is expected to rapidly deplete relatedness providing that the rate of outcrossing is sufficiently high.
Evidence of IBD between parasite samples has been observed to positively correlate with reductions in parasite transmission 11 , 101 , and bottlenecks in vector population size similarly induce inbreeding 29 , indicating the utility of this feature as a metric for assessing changes in transmission intensity and vector population size. In the fields of fisheries and conservation biology, relatedness prevalence has long been used as a tool for estimating the abundance of a given species, for example, through the technique of close kin mark recapture 161 , 162 . Although the life cycles of malaria parasites and anopheline mosquitoes differ in ways that require distinct interpretations of relatedness, the fundamental premise that related individuals are more likely to be co-sampled in smaller populations than larger ones makes this technique equally apt for both parasites and vectors. Relatedness inference has other applications that are beginning to be explored in the malaria context. For example, evidence of 100% relatedness associated with clones has been used to classify P. falciparum recurrences as either newly acquired or a product of treatment failure 163 , and evidence of 50% relatedness associated with outcrossed siblings has been used to help classify P. vivax recurrences as liver-stage relapses 164 . Finally, relatedness estimates can be subjected to downstream analyses, including various clustering algorithms and statistical models that can explicitly articulate hypotheses about malaria epidemiology 165 .
Conclusions and perspectives
The malaria community is fortunate to have amassed a wealth of genomic data resources and analytical methods to support applications, from fundamental malaria biology to applied malaria control strategies. Many studies have shown the value of these resources for identifying markers and mechanisms of drug and insecticide resistance and for complementing traditional epidemiological disease indicators such as case counts, incidence and travel history with genomic data, in order to inform relative rates of local transmission against case importation, connectivity between populations and efficacy of parasite and vector interventions.
As the research community will undoubtedly continue to make genomic data more practical to generate and use in the fight against malaria, the next problem to solve will be how to manage all of the distributed data generation resulting from enhanced parasite and vector surveillance efforts taking place in disease-endemic countries. Three key gaps remain in realizing the potential of genomic epidemiology for real-world malaria control and elimination strategy: first, local experts should be able to carry out data generation, analysis and interpretation without extensive support from highly specialized and well-resourced institutions; second, to be of practical value genomic information must be produced on operationally relevant timescales; and third, analysis across genomic data sets and integration with epidemiological metrics must be facilitated, by both encouraging interoperability between data sets and establishing networks between stakeholders.
Given current capacities for data generation and limitations in data computing, we anticipate that information from targeted approaches, such as PCR amplicon-based or MIP-based sequencing and SNP-based genotyping, will foreseeably form the basis of most data generation efforts within malaria-endemic countries as they remain viable to implement at scale in lower resource settings than WGS. Collaborating institutions can generate these data types to support countries and research partners and for research discovery purposes. In parallel, we anticipate that WGS data will continue to inform the understanding of global genetic diversity and dynamics on a genome-wide scale, and continuously improve the design strategy and data interpretation framework for targeted sequencing and genotyping approaches.
The potential value of using variation caused by recombination as a framework for interpreting malaria genomic data and informing disease intervention strategies, as opposed to using variation caused by mutation (as in the genomic epidemiology of measurably evolving pathogens) or variation in allele frequencies (as captured by π and F ST ), has already influenced the development of targeted sequencing approaches optimized for relatedness inference. For example, targeted resequencing efforts have begun to focus on closely clustered SNP variants that give rise to many diverse haplotypes 142 , so as to better inform relatedness inference 149 . Additional investment will be necessary at the level of analysis and visualization to further identify and exploit signals in malaria genomic data that are relevant to decisions in public health. Key to this effort will be methods to store, house and share data and to support analysis workflows across studies and surveillance efforts that generate epidemiological data 10 , 166 , 167 . In addition to the NCBI Sequence Read Archive and the European Nucleotide Archive, the malaria field has long benefited from genomic data repositories for parasites (PlasmoDB) 168 and vectors (VectorBase) 169 , which have recently merged into a common platform called VEuPathDB . These databases have provided invaluable curation of reference genome assemblies and annotations, integrated with functional and transcriptomic data to provide insight into the roles of genes. However, their primary audience has been researchers; the operationalization of genomic surveillance methods may require a distinct approach to the management, analysis and sharing of genomic data to serve stakeholders in the public health community.
Although there are many technical considerations to be addressed, perhaps the single biggest issue is data sharing. As a community we must aspire to a future of open data sharing for the benefit of global health while being sensitive to the complexities that prevent data from being shared and released. We must also support countries as they work to understand the risks and benefits of sharing genomic data, set policy for the future and be pragmatic about serving current needs in the meantime. With this in place, genomic variation data at the population level will be empowered to drive transformative impacts in global public health.
Baton, L. A. & Ranford-Cartwright, L. C. Spreading the seeds of million-murdering death: metamorphoses of malaria in the mosquito. Trends Parasitol. 21 , 573–580 (2005).
Article PubMed Google Scholar
Loy, D. E. et al. Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax . Int. J. Parasitol. 47 , 87–97 (2017).
Otto, T. D. et al. Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria. Nat. Microbiol. 3 , 687–697 (2018).
Article CAS PubMed PubMed Central Google Scholar
Kwiatkowski, D. P. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet. 77 , 171–192 (2005).
World Health Organization. World Malaria Report 2020 (WHO, 2020).
Molina-Cruz, A., Zilversmit, M. M., Neafsey, D. E., Hartl, D. L. & Barillas-Mury, C. Mosquito vectors and the globalization of Plasmodium falciparum malaria. Annu. Rev. Genet. 50 , 447–465 (2016).
Article CAS PubMed Google Scholar
Ranson, H. & Lissenden, N. Insecticide resistance in African anopheles mosquitoes: a worsening situation that needs urgent action to maintain malaria control. Trends Parasitol. 32 , 187–196 (2016).
Ouattara, A. et al. Molecular basis of allele-specific efficacy of a blood-stage malaria vaccine: vaccine development implications. J. Infect. Dis. 207 , 511–519 (2013).
Jacob, C. G. et al. Genetic surveillance in the Greater Mekong Subregion and South Asia to support malaria control and elimination. Preprint at medRxiv https://doi.org/10.1101/2020.07.23.20159624 (2020).
Article PubMed PubMed Central Google Scholar
Wesolowski, A. et al. Mapping malaria by combining parasite genomic and epidemiologic data. BMC Med. 16 , 190 (2018).
Daniels, R. F. et al. Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proc. Natl Acad. Sci. USA 112 , 7067–7072 (2015).
Lee, H. J. et al. Transcriptomic studies of malaria: a paradigm for investigation of systemic host-pathogen interactions. Microbiol. Mol. Biol. Rev. 82 , e00071-17 (2018).
Lee, M. C. S., Lindner, S. E., Lopez-Rubio, J.-J. & Llinás, M. Cutting back malaria: CRISPR/Cas9 genome editing of Plasmodium . Brief. Funct. Genomics 18 , 281–289 (2019).
Article PubMed PubMed Central CAS Google Scholar
Kariuki, S. N. & Williams, T. N. Human genetics and malaria resistance. Hum. Genet. 139 , 801–811 (2020).
Gardner, M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum . Nature 419 , 498–511 (2002). This paper describes the first reference genome sequence assembly for P. falciparum .
Dame, J. B. et al. Current status of the Plasmodium falciparum genome project. Mol. Biochem. Parasitol. 79 , 1–12 (1996).
Jeffares, D. C. et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum . Nat. Genet. 39 , 120–125 (2007).
Mu, J. et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat. Genet. 39 , 126–130 (2007).
Volkman, S. K. et al. A genome-wide map of diversity in Plasmodium falciparum . Nat. Genet. 39 , 113–119 (2007). Along with Jeffares et al. and Mu et al., these three co-published papers describe the first efforts to characterize genome-wide variation in P. falciparum , using Sanger di-deoxy shotgun and PCR-based sequencing. They describe fundamental population genomic features of the parasite, including variation in population diversity, global population structure and patterns of linkage disequilibrium .
Neafsey, D. E. et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum . Nat. Genet. 44 , 1046–1050 (2012).
Hupalo, D. N. et al. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax . Nat. Genet. 48 , 953–958 (2016).
Neafsey, D. E. et al. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol. 9 , R171 (2008).
Van Tyne, D. et al. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum . PLoS Genet. 7 , e1001383 (2011).
Kidgell, C. et al. A systematic map of genetic variation in Plasmodium falciparum . PLoS Pathog. 2 , e57 (2006).
Holt, R. A. et al. The genome sequence of the malaria mosquito Anopheles gambiae . Science 298 , 129–149 (2002). This paper describes the first reference genome assembly for A. gambiae , produced from the PEST colony that represented an admixture of A. gambiae sensu stricto and A. coluzzii .
Sinka, M. E. et al. The dominant anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic précis. Parasit. Vectors 3 , 117 (2010).
Martinez-Torres, D. et al. Molecular characterization of pyrethroid knockdown resistance ( kdr ) in the major malaria vector Anopheles gambiae s.s . Insect Mol. Biol. 7 , 179–184 (1998).
Weill, M. et al. A novel acetylcholinesterase gene in mosquitoes codes for the insecticide target and is non-homologous to the ace gene in Drosophila. Proc. Biol. Sci. 269 , 2007–2016 (2002).
Anopheles gambiae 1000 Genomes Consortium. Genetic diversity of the African malaria vector Anopheles gambiae . Nature 552 , 96–100 (2017). This work describes the first large-scale population genomic study of A. gambiae sensu stricto and A. coluzzii performed on WGS data, documenting signals of selection associated with insecticide resistance and examples of resistance alleles that crossed species boundaries through hybridization and introgression .
Article CAS Google Scholar
Lucas, E. R. et al. Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes. Genome Res. 29 , 1250–1261 (2019).
Coetzee, M. et al. Anopheles coluzzii and Anopheles amharicus , new members of the Anopheles gambiae complex. Zootaxa 3619 , 246–274 (2013).
Barrón, M. G. et al. A new species in the major malaria vector complex sheds light on reticulated species evolution. Sci. Rep. 9 , 14753 (2019).
Fontaine, M. C. et al. Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347 , 1258524 (2015).
Article PubMed CAS Google Scholar
Crawford, J. E. et al. Reticulate speciation and barriers to introgression in the Anopheles gambiae species complex. Genome Biol. Evol. 7 , 3116–3131 (2015).
Crawford, J. E. et al. Evolution of GOUNDRY, a cryptic subgroup of Anopheles gambiae s.l ., and its impact on susceptibility to Plasmodium infection. Mol. Ecol. 25 , 1494–1510 (2016).
Riehle, M. M. et al. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science 331 , 596–598 (2011).
Davidson, G. Insecticide resistance in Anopheles gambiae Giles. Nature 178 , 705–706 (1956).
Davidson, G. Insecticide resistance in Anopheles gambiae Giles: a case of simple Mendelian inheritance. Nature 178 , 863–864 (1956).
Davidson, G. & Jackson, C. E. Incipient speciation in Anopheles gambiae Giles. Bull. World Health Organ. 27 , 303–305 (1962).
CAS PubMed PubMed Central Google Scholar
Moreno, M. et al. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar. J. 9 , 127 (2010).
Carlton, J. M. et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax . Nature 455 , 757–763 (2008). This paper describes the first reference genome assembly for P. vivax , which exhibits a less extreme level of A/T nucleotide composition bias than P. falciparum .
Neafsey, D. E. et al. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347 , 1258522 (2015).
Ghurye, J. et al. A chromosome-scale assembly of the major African malaria vector Anopheles funestus . GigaScience 8 , giz063 (2019).
Marinotti, O. et al. The genome of Anopheles darlingi , the main neotropical malaria vector. Nucleic Acids Res. 41 , 7387–7400 (2013).
Carlton, J. M. et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii . Nature 419 , 512–519 (2002).
Manske, M. et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487 , 375–379 (2012). The first paper to describe the population genomics of a worldwide collection of P. falciparum isolates. It demonstrates the power of WGS data for identifying signals of natural selection and fine-scale population structure in parasite populations .
Pearson, R. D. et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax . Nat. Genet. 48 , 959–964 (2016).
Miotto, O. et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum . Nat. Genet. 47 , 226–234 (2015).
Clarkson, C. S., Temple, H. J. & Miles, A. The genomics of insecticide resistance: insights from recent studies in African malaria vectors. Curr. Opin. Insect Sci. 27 , 111–115 (2018).
Ghansah, A. et al. Monitoring parasite diversity for malaria elimination in sub-Saharan Africa. Science 345 , 1297–1298 (2014).
Lan, J. H. et al. Impact of three Illumina library construction methods on GC bias and HLA genotype calling. Hum. Immunol. 76 , 166–175 (2015).
Trager, W. & Jensen, J. B. Human malaria parasites in continuous culture. Science 193 , 673–675 (1976).
Claessens, A., Affara, M., Assefa, S. A., Kwiatkowski, D. P. & Conway, D. J. Culture adaptation of malaria parasites selects for convergent loss-of-function mutants. Sci. Rep. 7 , 41303 (2017).
Gunalan, K., Rowley, E. H. & Miller, L. H. A way forward for culturing Plasmodium vivax . Trends Parasitol. 36 , 512–519 (2020).
Venkatesan, M. et al. Using CF11 cellulose columns to inexpensively and effectively remove human DNA from Plasmodium falciparum -infected whole blood samples. Malar. J. 11 , 41 (2012).
Melnikov, A. et al. Hybrid selection for sequencing pathogen genomes from clinical samples. Genome Biol. 12 , R73 (2011).
Bright, A. T. et al. Whole genome sequencing analysis of Plasmodium vivax using whole genome capture. BMC Genomics 13 , 262 (2012).
Leichty, A. R. & Brisson, D. Selective whole genome amplification for resequencing target microbial species from complex natural samples. Genetics 198 , 473–481 (2014).
Oyola, S. O. et al. Whole genome sequencing of Plasmodium falciparum from dried blood spots using selective whole genome amplification. Malar. J. 15 , 597 (2016).
Cowell, A. N. et al. Selective whole-genome amplification is a robust method that enables scalable whole-genome sequencing of Plasmodium vivax from unprocessed clinical samples. mBio 8 , e02257 (2017).
Miotto, O. et al. Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea. PLoS Pathog. 16 , e1009133 (2020).
Mathieu, L. C. et al. Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance. eLife 9 , e51015 (2020).
Antonio-Nkondjio, C. & Simard, F. in Anopheles Mosquitoes: New Insights into Malaria Vectors (ed. Manguin, S.) (InTech, 2013).
St Laurent, B. et al. Behaviour and molecular identification of Anopheles malaria vectors in Jayapura district, Papua province, Indonesia. Malar. J. 15 , 192 (2016).
St Laurent, B. et al. Molecular characterization reveals diverse and unknown malaria vectors in the Western Kenyan Highlands. Am. J. Trop. Med. Hyg. 94 , 327–335 (2016).
Ruiz-Lopez, F. et al. DNA barcoding reveals both known and novel taxa in the albitarsis group ( Anopheles : Nyssorhynchus ) of neotropical malaria vectors. Parasit. Vectors 5 , 44 (2012).
Ahumada, M. L. et al. Spatial distributions of Anopheles species in relation to malaria incidence at 70 localities in the highly endemic Northwest and South Pacific coast regions of Colombia. Malar. J. 15 , 407 (2016).
Beebe, N. W., Russell, T., Burkot, T. R. & Cooper, R. D. Anopheles punctulatus group: evolution, distribution, and control. Annu. Rev. Entomol. 60 , 335–350 (2015).
Garg, S. et al. A haplotype-aware de novo assembly of related individuals using pedigree sequence graph. Bioinformatics 36 , 2385–2392 (2020).
Korlach, J. et al. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. GigaScience 6 , 1–16 (2017).
Kingan, S. B. et al. A high-quality de novo genome assembly from a single mosquito using PacBio sequencing. Genes 10 , 62 (2019).
Article PubMed Central CAS Google Scholar
Roper, C. et al. Intercontinental spread of pyrimethamine-resistant malaria. Science 305 , 1124 (2004).
Reeder, J. C. & Marshall, V. M. A simple method for typing Plasmodium falciparum merozoite surface antigens 1 and 2 (MSA-1 and MSA-2) using a dimorphic-form specific polymerase chain reaction. Mol. Biochem. Parasitol. 68 , 329–332 (1994).
Snounou, G. Genotyping of Plasmodium spp. Nested PCR. Methods Mol. Med. 72 , 103–116 (2002).
CAS PubMed Google Scholar
Daniels, R. et al. A general SNP-based molecular barcode for Plasmodium falciparum identification and tracking. Malar. J. 7 , 223 (2008). The first paper to describe a ‘molecular barcoding’ approach to genotyping parasites, in this case 24 biallelic SNPs selected on the basis of high minor allele frequency in Senegal and Thai P. falciparum populations. It demonstrates the utility of inexpensive genotyping data for diverse applications .
Baniecki, M. L. et al. Development of a single nucleotide polymorphism barcode to genotype Plasmodium vivax infections. PLoS Negl. Trop. Dis. 9 , e0003539 (2015).
Campino, S. et al. Population genetic analysis of Plasmodium falciparum parasites using a customized Illumina GoldenGate genotyping assay. PLoS ONE 6 , e20251 (2011).
Lee, Y., Marsden, C. D., Nieman, C. & Lanzaro, G. C. A new multiplex SNP genotyping assay for detecting hybridization and introgression between the M and S molecular forms of Anopheles gambiae . Mol. Ecol. Resour. 14 , 297–305 (2014).
Neafsey, D. E. et al. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science 330 , 514–517 (2010).
Bailey, J. A. et al. Use of massively parallel pyrosequencing to evaluate the diversity of and selection on Plasmodium falciparum csp T-cell epitopes in Lilongwe, Malawi. J. Infect. Dis. 206 , 580–587 (2012).
Aragam, N. R. et al. Diversity of T cell epitopes in Plasmodium falciparum circumsporozoite protein likely due to protein-protein interactions. PLoS ONE 8 , e62427 (2013).
Juliano, J. J. et al. Exposing malaria in-host diversity and estimating population diversity by capture-recapture using massively parallel pyrosequencing. Proc. Natl Acad. Sci. USA 107 , 20138–20143 (2010).
Gandhi, K. et al. Variation in the circumsporozoite protein of Plasmodium falciparum : vaccine development implications. PLoS ONE 9 , e101783 (2014).
Early, A. M. et al. Host-mediated selection impacts the diversity of Plasmodium falciparum antigens within infections. Nat. Commun. 9 , 1381 (2018).
Aydemir, O. et al. Drug-resistance and population structure of Plasmodium falciparum across the Democratic Republic of Congo using high-throughput molecular inversion probes. J. Infect. Dis. 218 , 946–955 (2018).
Shetty, A. C. et al. Genomic structure and diversity of Plasmodium falciparum in Southeast Asia reveal recent parasite migration patterns. Nat. Commun. 10 , 2665 (2019).
Auburn, S. et al. Genomic analysis of Plasmodium vivax in Southern Ethiopia reveals selective pressures in multiple parasite mechanisms. J. Infect. Dis. 220 , 1738–1749 (2019).
Amambua-Ngwa, A. et al. Major subpopulations of Plasmodium falciparum in sub-Saharan Africa. Science 365 , 813–816 (2019). This paper describes efforts by the PDNA to characterize African P. falciparum genomic diversity using WGS data. The authors describe signals of natural selection and population structure within the diverse parasite population on this continent .
Miotto, O. et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet. 45 , 648–655 (2013).
Haldar, K., Bhattacharjee, S. & Safeukui, I. Drug resistance in Plasmodium . Nat. Rev. Microbiol. 16 , 156–170 (2018).
Amato, R. et al. Origins of the current outbreak of multidrug-resistant malaria in southeast Asia: a retrospective genetic study. Lancet Infect. Dis. 18 , 337–345 (2018). A detailed genomic portrait of the emergence and spread of mutations associated with resistance to artemisinin and co-administered partner drugs in the GMS. It demonstrates the rich value of geographically comprehensive parasite genomic data collected longitudinally, for example, in providing the insight that pfkelch13 artemisinin resistance mutations arose at least 30 times independently .
Cheeseman, I. H. et al. A major genome region underlying artemisinin resistance in malaria. Science 336 , 79–82 (2012).
World Health Organization. A framework for malaria elimination (WHO, 2017).
Fidock, D. A. et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6 , 861–871 (2000).
Wellems, T. E. Transporter of a malaria catastrophe. Nat. Med. 10 , 1169–1171 (2004).
Peterson, D. S., Walliker, D. & Wellems, T. E. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl Acad. Sci. USA 85 , 9114–9118 (1988).
Noedl, H. et al. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 359 , 2619–2620 (2008).
Dondorp, A. M. et al. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361 , 455–467 (2009).
Ariey, F. et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505 , 50–55 (2014).
Straimer, J. et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347 , 428–431 (2015).
Cerqueira, G. C. et al. Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol. 18 , 78 (2017).
Li, X. et al. Genetic mapping of fitness determinants across the malaria parasite Plasmodium falciparum life cycle. PLoS Genet. 15 , e1008453 (2019).
Takala-Harrison, S. et al. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J. Infect. Dis. 211 , 670–679 (2015).
Anderson, T. J. C. et al. Population parameters underlying an ongoing soft sweep in southeast Asian malaria parasites. Mol. Biol. Evol. 34 , 131–144 (2017).
Ansbro, M. R. et al. Development of copy number assays for detection and surveillance of piperaquine resistance associated plasmepsin 2/3 copy number variation in Plasmodium falciparum . Malar. J. 19 , 181 (2020).
Chenet, S. M. et al. Independent emergence of the Plasmodium falciparum kelch propeller domain mutant allele C580Y in Guyana. J. Infect. Dis. 213 , 1472–1475 (2016).
Uwimana, A. et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 26 , 1602–1608 (2020).
Taylor, S. M. et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J. Infect. Dis. 211 , 680–688 (2015).
Tessema, S. K. et al. Applying next-generation sequencing to track falciparum malaria in sub-Saharan Africa. Malar. J. 18 , 268 (2019).
Meshnick, S. Artemisinin resistance in Southeast Asia. Clin. Infect. Dis. 63 , 1527 (2016).
Hastings, I. M., Kay, K. & Hodel, E. M. The importance of scientific debate in the identification, containment, and control of artemisinin resistance. Clin. Infect. Dis. 63 , 1527–1528 (2016).
Phyo, A. P. et al. Reply to Meshnick and Hastings et al. Clin. Infect. Dis. 63 , 1528–1529 (2016).
Ranson, H. Current and future prospects for preventing malaria transmission via the use of insecticides. Cold Spring Harb. Perspect. Med. 7 , a026823 (2017).
Bhatt, S. et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 526 , 207–211 (2015).
Weill, M. et al. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol. Biol. 13 , 1–7 (2004).
Ranson, H. et al. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae . Biochem. J. 359 , 295–304 (2001).
Jones, C. M. et al. The dynamics of pyrethroid resistance in Anopheles arabiensis from Zanzibar and an assessment of the underlying genetic basis. Parasit. Vectors 6 , 343 (2013).
Ingham, V. A. et al. A sensory appendage protein protects malaria vectors from pyrethroids. Nature 577 , 376–380 (2020).
Bailey, J. A. et al. Microarray analyses reveal strain-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 variants following natural infection and vaccination. Sci. Rep. 10 , 3952 (2020).
Ouattara, A. et al. Designing malaria vaccines to circumvent antigen variability. Vaccine 33 , 7506–7512 (2015).
Thera, M. A. et al. A field trial to assess a blood-stage malaria vaccine. N. Engl. J. Med. 365 , 1004–1013 (2011).
Neafsey, D. E. et al. Genetic diversity and protective efficacy of the RTS,S/AS01 malaria vaccine. N. Engl. J. Med. 373 , 2025–2037 (2015).
Amambua-Ngwa, A. et al. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet. 8 , e1002992 (2012).
Sauerwein, R. W. & Bousema, T. Transmission blocking malaria vaccines: assays and candidates in clinical development. Vaccine 33 , 7476–7482 (2015).
Bustamante, L. Y. et al. A full-length recombinant Plasmodium falciparum PfRH5 protein induces inhibitory antibodies that are effective across common PfRH5 genetic variants. Vaccine 31 , 373–379 (2013).
Douglas, A. D. et al. The blood-stage malaria antigen PfRH5 is susceptible to vaccine-inducible cross-strain neutralizing antibody. Nat. Commun. 2 , 601 (2011).
Tan, J. et al. A public antibody lineage that potently inhibits malaria infection through dual binding to the circumsporozoite protein. Nat. Med. 24 , 401–407 (2018).
Kisalu, N. K. et al. A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite. Nat. Med. 24 , 408–416 (2018).
Smith-Aguasca, R. et al. Mosquitoes as a feasible sentinel group for anti-malarial resistance surveillance by next generation sequencing of Plasmodium falciparum . Malar. J. 18 , 351 (2019).
Sumner, K. M. et al. Genotyping cognate Plasmodium falciparum in humans and mosquitoes to estimate onward transmission of asymptomatic infections. Nat. Commun. 12 , 909 (2021).
Nkhoma, S. et al. Population genetic correlates of declining transmission in a human pathogen. Mol. Ecol. 22 , 273–285 (2013). This manuscript performs a rigorous evaluation of multiple population genetic signatures of declining transmission along the Thailand–Myanmar border, finding that the proportion of complex infections provides the strongest signal .
Galinsky, K. et al. COIL: a methodology for evaluating malarial complexity of infection using likelihood from single nucleotide polymorphism data. Malar. J. 14 , 4 (2015).
Medica, D. L. & Sinnis, P. Quantitative dynamics of Plasmodium yoelii sporozoite transmission by infected anopheline mosquitoes. Infect. Immun. 73 , 4363–4369 (2005).
Amino, R. et al. Quantitative imaging of Plasmodium transmission from mosquito to mammal. Nat. Med. 12 , 220–224 (2006).
Wong, W., Wenger, E. A., Hartl, D. L. & Wirth, D. F. Modeling the genetic relatedness of Plasmodium falciparum parasites following meiotic recombination and cotransmission. PLoS Comput. Biol. 14 , e1005923 (2018).
Nkhoma, S. C. et al. Close kinship within multiple-genotype malaria parasite infections. Proc. Biol. Sci. 279 , 2589–2598 (2012).
PubMed PubMed Central Google Scholar
Nair, S. et al. Single-cell genomics for dissection of complex malaria infections. Genome Res. 24 , 1028–1038 (2014).
Nkhoma, S. C. et al. Co-transmission of related malaria parasite lineages shapes within-host parasite diversity. Cell Host Microbe 27 , 93–103.e4 (2020). This study employs single-cell WGS data of 485 parasites from 15 clinical infection samples to demonstrate strong evidence for serial co-transmission of genetically distinct malaria parasites in Malawi, a high-transmission setting. Prior to this work, complex infections in such a setting were assumed to be generated primarily through superinfection rather than co-transmission .
Dalmat, R., Naughton, B., Kwan-Gett, T. S., Slyker, J. & Stuckey, E. M. Use cases for genetic epidemiology in malaria elimination. Malar. J. 18 , 163 (2019).
Noviyanti, R. et al. Implementing parasite genotyping into national surveillance frameworks: feedback from control programmes and researchers in the Asia-Pacific region. Malar. J. 19 , 271 (2020).
Apinjoh, T. O., Ouattara, A., Titanji, V. P. K., Djimde, A. & Amambua-Ngwa, A. Genetic diversity and drug resistance surveillance of Plasmodium falciparum for malaria elimination: is there an ideal tool for resource-limited sub-Saharan Africa? Malar. J. 18 , 217 (2019).
Tessema, S. K. et al. Sensitive, highly multiplexed sequencing of microhaplotypes from the Plasmodium falciparum heterozygome. J. Infect. Dis. https://doi.org/10.1093/infdis/jiaa527 (2020). In this paper, the authors describe an amplicon sequencing panel for P. falciparum designed with the express purpose of profiling highly heterozygous microhaplotypes for the purpose of IBD inference .
Article PubMed Central Google Scholar
McDew-White, M. et al. Mode and tempo of microsatellite length change in a malaria parasite mutation accumulation experiment. Genome Biol. Evol. 11 , 1971–1985 (2019).
Nei, M. & Li, W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA 76 , 5269–5273 (1979).
Nei, M. Molecular Evolutionary Genetics (Columbia Univ. Press, 1987).
Balding, D. J. Likelihood-based inference for genetic correlation coefficients. Theor. Popul. Biol. 63 , 221–230 (2003).
Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145 , 1219–1228 (1997).
Wright, S. Coefficients of inbreeding and relationship. Am. Nat. 56 , 330–338 (1922).
Article Google Scholar
Taylor, A. R., Jacob, P. E., Neafsey, D. E. & Buckee, C. O. Estimating relatedness between malaria parasites. Genetics 212 , 1337–1351 (2019).
Weir, B. S., Anderson, A. D. & Hepler, A. B. Genetic relatedness analysis: modern data and new challenges. Nat. Rev. Genet. 7 , 771–780 (2006).
Thompson, E. A. Identity by descent: variation in meiosis, across genomes, and in populations. Genetics 194 , 301–326 (2013).
Henden, L., Lee, S., Mueller, I., Barry, A. & Bahlo, M. Identity-by-descent analyses for measuring population dynamics and selection in recombining pathogens. PLoS Genet. 14 , e1007279 (2018).
Speed, D. & Balding, D. J. Relatedness in the post-genomic era: is it still useful? Nat. Rev. Genet. 16 , 33–44 (2015).
Schaffner, S. F., Taylor, A. R., Wong, W., Wirth, D. F. & Neafsey, D. E. hmmIBD: software to infer pairwise identity by descent between haploid genotypes. Malar. J. 17 , 196 (2018).
Taylor, A. R., Echeverry, D. F., Anderson, T. J. C., Neafsey, D. E. & Buckee, C. O. Identity-by-descent with uncertainty characterises connectivity of Plasmodium falciparum populations on the Colombian-Pacific coast. PLoS Genet. 16 , e1009101 (2020).
Biek, R., Pybus, O. G., Lloyd-Smith, J. O. & Didelot, X. Measurably evolving pathogens in the genomic era. Trends Ecol. Evol. 30 , 306–313 (2015).
Lemieux, J. Genomic Analysis of Evolution in Plasmodium falciparum and Babesia microti . Thesis, Harvard Medical School (2015).
Hamilton, W. L. et al. Extreme mutation bias and high AT content in Plasmodium falciparum . Nucleic Acids Res. 45 , 1889–1901 (2017).
Redmond, S. N. et al. De novo mutations resolve disease transmission pathways in clonal malaria. Mol. Biol. Evol. 35 , 1678–1689 (2018).
Taylor, A. R. et al. Quantifying connectivity between local Plasmodium falciparum malaria parasite populations using identity by descent. PLoS Genet. 13 , e1007065 (2017). This manuscript, which analyses P. falciparum 93-SNP barcode and WGS data from the Thailand–Myanmar border, is the first to demonstrate that IBD-based relatedness estimates can be a useful tool for resolving parasite population connectivity over a geographic scale of less than 100 km and that they can be computed using SNP barcode as well as WGS data .
Skaug, H. J. Allele-sharing methods for estimation of population size. Biometrics 57 , 750–756 (2001).
Bravington, M., Skaug, H. & Anderson, E. Close-kin mark-recapture. Stat. Sci. 31 , 259–274 (2016).
Plucinski, M. M., Morton, L., Bushman, M., Dimbu, P. R. & Udhayakumar, V. Robust algorithm for systematic classification of malaria late treatment failures as recrudescence or reinfection using microsatellite genotyping. Antimicrob. Agents Chemother. 59 , 6096–6100 (2015).
Taylor, A. R. et al. Resolving the cause of recurrent Plasmodium vivax malaria probabilistically. Nat. Commun. 10 , 5595 (2019).
Watson, J. A. et al. A cautionary note on the use of unsupervised machine learning algorithms to characterise malaria parasite population structure from genetic distance matrices. PLoS Genet. 16 , e1009037 (2020).
Tessema, S. et al. Using parasite genetic and human mobility data to infer local and cross-border malaria connectivity in Southern Africa. eLife 8 , e43510 (2019).
Chang, H.-H. et al. Mapping imported malaria in Bangladesh using parasite genetic and human mobility data. eLife 8 , e43481 (2019).
Bahl, A. et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 31 , 212–215 (2003).
Lawson, D. et al. VectorBase: a home for invertebrate vectors of human pathogens. Nucleic Acids Res. 35 , D503–D505 (2007).
Pearson, R. D., Amato, R., Kwiatkowski, D. P. & MalariaGEN Plasmodium falciparum Community Project. An open dataset of Plasmodium falciparum genome variation in 7,000 worldwide samples. Preprint at bioRxiv https://doi.org/10.1101/824730 (2019).
Turner, T. L., Hahn, M. W. & Nuzhdin, S. V. Genomic islands of speciation in Anopheles gambiae . PLoS Biol. 3 , e285 (2005).
Galen, S. C. et al. The polyphyly of Plasmodium : comprehensive phylogenetic analyses of the malaria parasites (order Haemosporida) reveal widespread taxonomic conflict. R. Soc. Open Sci. 5 , 171780 (2018).
Eick, G. N., Jacobs, D. S. & Matthee, C. A. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (chiroptera). Mol. Biol. Evol. 22 , 1869–1886 (2005).
Foster, P. G. et al. Phylogeny of Anophelinae using mitochondrial protein coding genes. R. Soc. Open Sci. 4 , 170758 (2017).
Duffy, P. E. & Patrick Gorres, J. Malaria vaccines since 2000: progress, priorities, products. NPJ Vaccines 5 , 1–9 (2020).
Richard, G.-F., Kerrest, A. & Dujon, B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 72 , 686–727 (2008).
Imwong, M. et al. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect. Dis. 17 , 491–497 (2017).
Imwong, M., Hien, T. T., Thuy-Nhien, N. T., Dondorp, A. M. & White, N. J. Spread of a single multidrug resistant malaria parasite lineage (PfPailin) to Vietnam. Lancet Infect. Dis. 17 , 1022–1023 (2017).
Volkman, S. K., Neafsey, D. E., Schaffner, S. F., Park, D. J. & Wirth, D. F. Harnessing genomics and genome biology to understand malaria biology. Nat. Rev. Genet. 13 , 315–328 (2012).
Rao, P. N. et al. A method for amplicon deep sequencing of drug resistance genes in Plasmodium falciparum clinical isolates from India. J. Clin. Microbiol. 54 , 1500–1511 (2016).
Ngondi, J. M. et al. Surveillance for sulfadoxine-pyrimethamine resistant malaria parasites in the Lake and Southern Zones, Tanzania, using pooling and next-generation sequencing. Malar. J. 16 , 236 (2017).
Verity, R. et al. The impact of antimalarial resistance on the genetic structure of Plasmodium falciparum in the DRC. Nat. Commun. 11 , 2107 (2020).
Download references
Acknowledgements
The authors thank R. Amato for his thoughtful perspective in planning this Review. They thank A. Early for providing the calculations underlying Fig. 3.
Author information
These authors contributed equally: Daniel E. Neafsey, Aimee R. Taylor.
Authors and Affiliations
Department of Immunology and Infectious Diseases, Harvard T.H. Chan School of Public Health, Boston, MA, USA
Daniel E. Neafsey
Infectious Disease and Microbiome Program, Broad Institute, Cambridge, MA, USA
Daniel E. Neafsey, Aimee R. Taylor & Bronwyn L. MacInnis
Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA, USA
Aimee R. Taylor
You can also search for this author in PubMed Google Scholar
Contributions
D.N. and B.M. wrote the first draft of the manuscript. D.N. and A.T. designed the figures. All authors edited and reviewed the manuscript.
Corresponding authors
Correspondence to Daniel E. Neafsey or Bronwyn L. MacInnis .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Peer review information.
Nature Reviews Genetics thanks J. Carlton, J. Kissinger and E. Winzeler for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
ClinicalTrials.gov: https://clinicaltrials.gov/
Pf3k resource: https://www.malariagen.net/projects/pf3k
VEuPathDB: https://veupathdb.org/
Units of a genome assembly constructed from overlapping sequencing reads but representing separate homologous haplotypes from the same genomic region in a diploid sequencing template.
Deviation from a single randomly mating population owing to demographic, geographical and/or evolutionary factors.
(COI). The number of distinct genomic sequences that describe malaria parasites simultaneously infecting a vertebrate host.
The separate transfer of genetically distinct parasites to a vertebrate host over multiple mosquito bites.
The simultaneous transfer of genetically distinct parasites to a vertebrate host through a single mosquito bite.
Describes the mean pairwise genetic difference rate between isolates within a sample; often used as a proxy of effective population size, with larger populations having higher diversity.
( F ST ). A commonly used statistic in population genetics, describing genetic differentiation between populations, which may be defined in various ways, including as a function of the variance in allele frequencies between populations.
(IBD). A descriptor of individuals, chromosomal segments or alleles that are descended from a recent common ancestor; identity by descent is a latent state the probability of which can be inferred under a statistical model or deduced from a pedigree.
(IBS). A descriptor of individuals, chromosomal segments or alleles that share the same genetic sequence; identity by state is a state that can be observed in genetic data.
Rights and permissions
Reprints and permissions
About this article
Cite this article.
Neafsey, D.E., Taylor, A.R. & MacInnis, B.L. Advances and opportunities in malaria population genomics. Nat Rev Genet 22 , 502–517 (2021). https://doi.org/10.1038/s41576-021-00349-5
Download citation
Accepted : 03 March 2021
Published : 08 April 2021
Issue Date : August 2021
DOI : https://doi.org/10.1038/s41576-021-00349-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Plasmodium falciparum genetic diversity and multiplicity of infection based on msp-1, msp-2, glurp and microsatellite genetic markers in sub-saharan africa: a systematic review and meta-analysis.
- Alex Mwesigwa
- Pauline Byakika-Kibwika
Malaria Journal (2024)
Transcriptomic analysis of Anopheles gambiae from Benin reveals overexpression of salivary and cuticular proteins associated with cross-resistance to pyrethroids and organophosphates
- Helga Saizonou
- Lucy Mackenzie Impoinvil
- Luc S. Djogbénou
BMC Genomics (2024)
Tracing the origins of Plasmodium vivax resurgence after malaria elimination on Aneityum Island in Vanuatu
- Chim W. Chan
- Akira Kaneko
Communications Medicine (2024)
Genomics reveals heterogeneous Plasmodium falciparum transmission and selection signals in Zambia
- Abebe A. Fola
- Giovanna Carpi
Evaluating the performance of Plasmodium falciparum genetic metrics for inferring National Malaria Control Programme reported incidence in Senegal
- Wesley Wong
- Stephen F. Schaffner
- Sarah K. Volkman
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

IMAGES
VIDEO
COMMENTS
Page 1 of 164. Sort by. A five years malaria surveillance data analysis of North Shewa zone, Amhara region, Ethiopia: July 2018 to June 2023. Malaria is a critical public health concern in Ethiopia, with significant socioeconomic consequences.
Methods: In this paper, the international scientific publication on malaria and P. vivax has been analyzed using the Scopus database to try to define global trends in this field of study. Results : It has been shown that events such as the emergence of resistance to certain drugs can break a trend.
Primate malaria. Cross journal collections - closed. Contribution of climate change to the spread of infectious diseases. Human migration, conflict and infectious diseases. Spatial inequality, infectious diseases and disease control. 2020. Thematic series Malaria and COVID-19. Thematic series Malaria Elimination Demonstration Project, Mandla ...
Malaria is a mosquito-borne disease that is caused by Plasmodium parasites. Patients with malaria experience flu-like symptoms and, in severe cases, the disease can progress to neurological...
collection. Collection 25 April 2022. Malaria research toward disease elimination. Malaria is a life-threatening febrile illness caused by Plasmodium spp. parasites that are transmitted to...
Published February 1, 2023. N Engl J Med 2023;388: e9. DOI: 10.1056/NEJMp2216703. VOL. 388 NO. 5. In this documentary video from the New England Journal of Medicine, physicians and scientists from...
52 Citations. 19 Altmetric. Metrics. Abstract. Almost 20 years have passed since the first reference genome assemblies were published for Plasmodium falciparum, the deadliest malaria parasite,...
This phase 2 trial showed that a single subcutaneous dose of L9LS at the highest dose tested (300 mg) provided protective efficacy of 69.9% against P. falciparum infection and 77.4% against...
Learn more about Research Topics. Explores all aspects of malaria from transmission to resistance, and treatment to prevention, to help deliver effective solutions and strategies to control and eliminate malaria.
1. Introduction. Malaria affected an estimated 219 million people causing 435,000 deaths in 2017 globally. This burden of morbidity and mortality is a result of more than a century of global effort and research aimed at improving the prevention, diagnosis, and treatment of malaria [ 1 ].