Society Homepage About Public Health Policy Contact
Data-driven hypothesis generation in clinical research: what we learned from a human subject study, article sidebar.


Submit your own article
Register as an author to reserve your spot in the next issue of the Medical Research Archives.
Join the Society
The European Society of Medicine is more than a professional association. We are a community. Our members work in countries across the globe, yet are united by a common goal: to promote health and health equity, around the world.
Join Europe’s leading medical society and discover the many advantages of membership, including free article publication.
Main Article Content
Hypothesis generation is an early and critical step in any hypothesis-driven clinical research project. Because it is not yet a well-understood cognitive process, the need to improve the process goes unrecognized. Without an impactful hypothesis, the significance of any research project can be questionable, regardless of the rigor or diligence applied in other steps of the study, e.g., study design, data collection, and result analysis. In this perspective article, the authors provide a literature review on the following topics first: scientific thinking, reasoning, medical reasoning, literature-based discovery, and a field study to explore scientific thinking and discovery. Over the years, scientific thinking has shown excellent progress in cognitive science and its applied areas: education, medicine, and biomedical research. However, a review of the literature reveals the lack of original studies on hypothesis generation in clinical research. The authors then summarize their first human participant study exploring data-driven hypothesis generation by clinical researchers in a simulated setting. The results indicate that a secondary data analytical tool, VIADS—a visual interactive analytic tool for filtering, summarizing, and visualizing large health data sets coded with hierarchical terminologies, can shorten the time participants need, on average, to generate a hypothesis and also requires fewer cognitive events to generate each hypothesis. As a counterpoint, this exploration also indicates that the quality ratings of the hypotheses thus generated carry significantly lower ratings for feasibility when applying VIADS. Despite its small scale, the study confirmed the feasibility of conducting a human participant study directly to explore the hypothesis generation process in clinical research. This study provides supporting evidence to conduct a larger-scale study with a specifically designed tool to facilitate the hypothesis-generation process among inexperienced clinical researchers. A larger study could provide generalizable evidence, which in turn can potentially improve clinical research productivity and overall clinical research enterprise.
Article Details
The Medical Research Archives grants authors the right to publish and reproduce the unrevised contribution in whole or in part at any time and in any form for any scholarly non-commercial purpose with the condition that all publications of the contribution include a full citation to the journal as published by the Medical Research Archives .
An official website of the United States government
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock Locked padlock icon ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.
- Publications
- Account settings
- Advanced Search
- Journal List
Hypothesis-generating research and predictive medicine
Leslie g biesecker.
- Author information
- Copyright and License information
Corresponding author E-mail [email protected]
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see http://genome.cshlp.org/site/misc/terms.xhtml ). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 3.0 Unported), as described at http://creativecommons.org/licenses/by-nc/3.0/ .
Genomics has profoundly changed biology by scaling data acquisition, which has provided researchers with the opportunity to interrogate biology in novel and creative ways. No longer constrained by low-throughput assays, researchers have developed hypothesis-generating approaches to understand the molecular basis of nature—both normal and pathological. The paradigm of hypothesis-generating research does not replace or undermine hypothesis-testing modes of research; instead, it complements them and has facilitated discoveries that may not have been possible with hypothesis-testing research. The hypothesis-generating mode of research has been primarily practiced in basic science but has recently been extended to clinical-translational work as well. Just as in basic science, this approach to research can facilitate insights into human health and disease mechanisms and provide the crucially needed data set of the full spectrum of genotype–phenotype correlations. Finally, the paradigm of hypothesis-generating research is conceptually similar to the underpinning of predictive genomic medicine, which has the potential to shift medicine from a primarily population- or cohort-based activity to one that instead uses individual susceptibility, prognostic, and pharmacogenetic profiles to maximize the efficacy and minimize the iatrogenic effects of medical interventions.
The goal of this article is to describe how recent technological changes provide opportunities to undertake novel approaches to biomedical research and to practice genomic preventive medicine. Massively parallel sequencing is the primary technology that will be addressed here ( Mardis 2008 ), but the principles apply to many other technologies, such as proteomics, metabolomics, transcriptomics, etc. Readers of this journal are well aware of the precipitous fall of sequencing costs over the last several decades. The consequence of this fall is that we are no longer in a scientific and medical world where the throughput (and the costs) of testing is the key limiting factor around which these enterprises are organized. Once one is released from this limiting factor, one may ask whether these enterprises should be reorganized. Here I outline the principles of how these enterprises are organized, show how high-throughput biology can allow alternative organizations of these enterprises to be considered, and show how biology and medicine are in many ways similar. The discussion includes three categories of enterprises: basic research, clinical research, and medical practice.

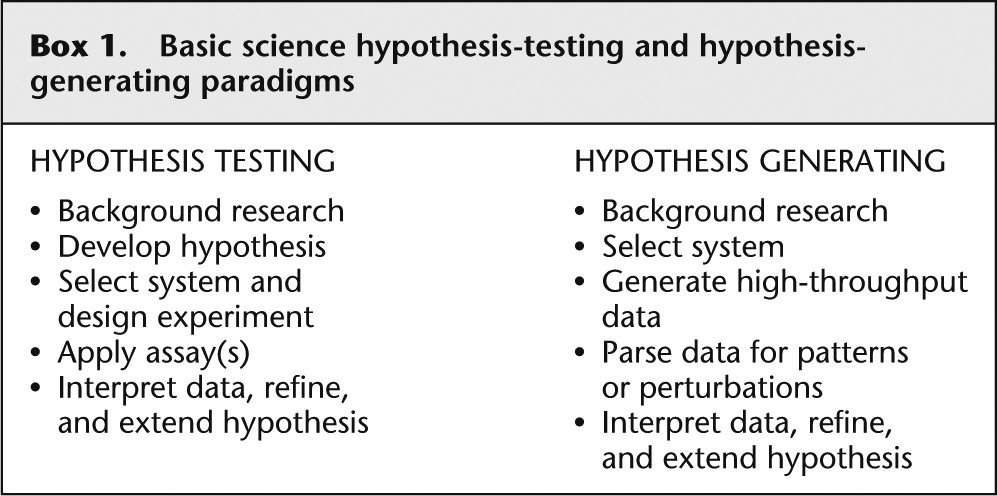
The basic science hypothesis-testing paradigm
The classical paradigm for basic biological research has been to develop a specific hypothesis that can be tested by the application of a prospectively defined experiment (see Box 1 ). I suggest that one of the major (although not the only) factors that led to the development of this paradigm is that experimental design was limited by the throughput of available assays. This low throughput mandated that the scientific question had to be focused narrowly to make the question tractable. However, the paradigm can be questioned if the scientist has the ability to assay every potential attribute of a given type (e.g., all genes). If the hypothesis is only needed to select the assay, one does not need a hypothesis to apply a technology that assays all attributes. In the case of sequencing, the radical increase in throughput can release scientists from the constraint of the specific hypothesis because it has allowed them to interrogate essentially all genotypes in a genome in a single assay. This capability facilitates fundamental biological discoveries that were impossible or impractical with a hypothesis-testing mode of scientific inquiry. Examples of this approach are well demonstrated by several discoveries that followed the sequencing of a number of genomes. An example was the discovery that the human gene count was just over 20,000 ( International Human Genome Sequencing Consortium 2004 ), much lower than prior estimates. This result, although it was much debated and anticipated, was not a hypothesis that drove the human genome project, but nonetheless was surprising and led to insights into the nuances of gene regulation and transcriptional isoforms to explain the complexity of the human organism. The availability of whole genome sequence data from multiple species facilitated analyses of conservation. While it was expected that protein-coding regions, and to a lesser extent promoters and 5′- and 3′-untranslated regions of genes, would exhibit recognizable sequence conservation, it was unexpected that an even larger fraction of the genomes outside of genes are highly conserved ( Mouse Genome Sequencing Consortium 2002 ). This surprising and unanticipated discovery has spawned a novel field of scientific inquiry to determine the functional roles of these elements, which are undoubtedly important in physiology and pathophysiology. These discoveries demonstrate the power of hypothesis-generating basic research to illuminate important biological principles.
Basic science hypothesis-testing and hypothesis-generating paradigms
Clinical and translational research
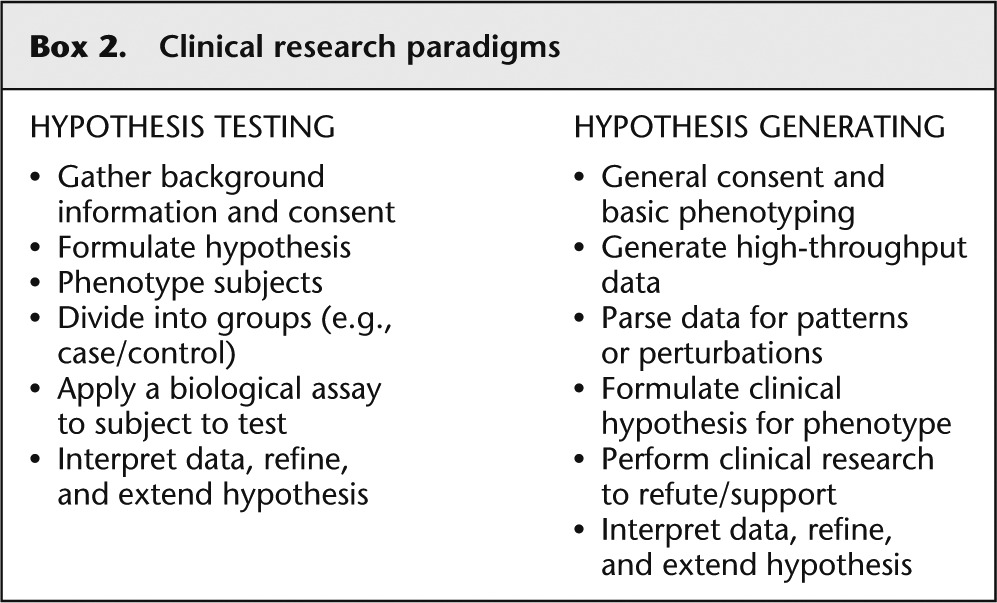
The approach to clinical research grew out of the basic science paradigm as described above. The first few steps of selecting a scientific problem and developing a hypothesis are similar, with the additional step ( Box 2 ) of rigorously defining a phenotype and then carefully selecting research participants with and without that trait. As in the basic science paradigm, the hypothesis is tested by the application of a specific assay to the cases and controls. Again, this paradigm has been incredibly fruitful and should not be abandoned, but the hypothesis-generating approach can be used here as well. In this approach, a cohort of participants is consented, basic information is gathered on their health, and then a high-throughput assay, such as genome or exome sequencing, is applied to all of the participants. Again, because the assay tests all such attributes, the research design does not necessitate a priori selections of phenotypes and genes to be interrogated. Then, the researcher can examine the sequence data set for patterns and perturbations, form hypotheses about how such perturbations might affect the phenotype of the participants, and test that hypothesis with a clinical research evaluation. This approach has been used with data from genome-wide copy number assessments (array CGH and SNP arrays), but sequencing takes it to a higher level of interrogation and provides innumerable variants with which to work.
Clinical research paradigms
An example of this type of sequence-based hypothesis-generating clinical research started with a collaborative project in which we showed that mutations in the gene ACSF3 caused the biochemical phenotype of combined malonic and methylmalonic acidemia ( Sloan et al. 2011 ). At that time, the disorder was believed to be a classic pediatric, autosomal-recessive severe metabolic disorder with decompensation and sometimes death. We then queried the ClinSeq cohort ( Biesecker et al. 2009 ) to assess the carrier frequency, to estimate the population frequency of this rare disorder. Because ClinSeq is a cohort of adults with a range of atherosclerosis severity, we reasoned that this would serve as a control population for an unbiased estimate of ACSF3 heterozygote mutant alleles. Surprisingly, we identified a ClinSeq participant who was homozygous for one of the mutations identified in the children with the typical phenotype. Indeed, one potential interpretation of the data would be that the variant is, in fact, benign and was erroneously concluded to be pathogenic, based on finding it in a child with the typical phenotype. It has been shown that this error is common, with up to 20% of variants listed in databases as pathogenic actually being benign ( Bell et al. 2011 ). Further clinical research on this participant led to the surprising result that she had severely abnormal blood and urine levels of malonic and methylmalonic acid ( Sloan et al. 2011 ). This novel approach to translational research was a powerful confirmation that the mutation was indeed pathogenic, but there was another, even more important conclusion. We had conceptualized the disease completely incorrectly. Instead of being only a severe, pediatric metabolic disorder, it was instead a disorder with a wide phenotypic spectrum in which one component of the disease is a metabolic perturbation and another component is a susceptibility to severe decompensation and strokes. This research indeed raises many questions about the natural history of the disorder, whether the pediatric decompensation phenotype is attributable to modifiers, what the appropriate management of such an adult would be, etc.
Irrespective of these limitations, the understanding of the disease has markedly advanced, and the key to understanding the broader spectrum of this disease was the hypothesis-generating approach enabled by the massively parallel sequence data and the ability to phenotype patients iteratively from ClinSeq. The iterative phenotyping was essential because we could not have anticipated when the patients were originally ascertained that we would need to assay malonic and methylmalonic acid. Nor did we recognize prospectively that we should be evaluating apparently healthy patients in their seventh decade for this phenotype. Indeed, it is impossible to evaluate patients for all potential phenotypes prospectively, and it is essential to minimize ascertainment bias for patient recruitment in order to allow the discovery of the full spectrum of phenotypes associated with genomic variations. This latter issue has become a critical challenge for implementing predictive medicine, as described below.
Predictive genomic medicine in practice
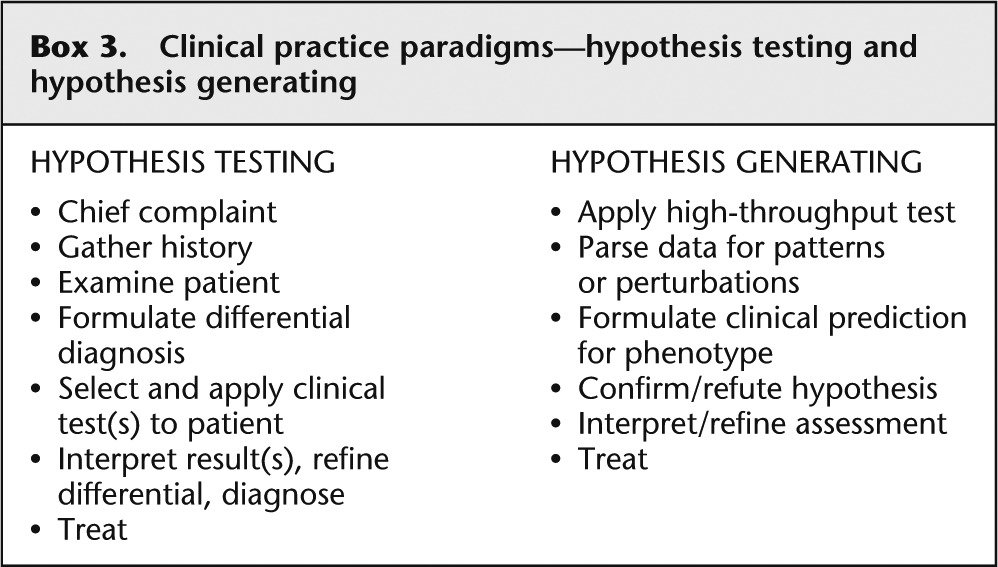
The principles of scientific inquiry are parallel to the processes of clinical diagnosis ( Box 3 ). In the classic, hypothesis-testing paradigm, clinicians gather background information including chief complaint, 2 medical and family history, and physical examination, and use these data to formulate the differential diagnosis, which is a set of potential medical diagnoses that could explain the patient's signs and symptoms. Then, the clinician selects, among the myriad of tests (imaging, biochemical, genetic, physiologic, etc.), a few tests, the results of which should distinguish among (or possibly exclude entirely) the disorders on the differential diagnosis. Like the scientist, the physician must act as a test selector, because each of the tests is low throughput, time consuming, and expensive.
Clinical practice paradigms—hypothesis testing and hypothesis generating
As in the basic and translational research discussion above, the question could be raised as to whether the differential diagnostic paradigm is necessary for genetic disorders. Indeed, the availability of clinical genome and exome sequencing heralds an era when the test could be ordered relatively early in the diagnostic process, with the clinician serving in a more interpretative role, rather than as a test selector ( Hennekam and Biesecker 2012 ). This approach has already been adopted for copy number variation, because whole genome array CGH- or SNP-based approaches have mostly displaced more specific single-gene or single-locus assays and standard chromosome analyses ( Miller et al. 2010 ). But the paradigm can be taken beyond hypothesis-generating clinical diagnosis into predictive medicine. One can now begin to envision how whole genome approaches could be used to assess risks prospectively for susceptibility to late-onset disorders or occult or subclinical disorders. The heritable cancer susceptibility syndromes are a good example of this. The current clinical approach is to order a specific gene test if a patient presents with a personal history of an atypical or early-onset form of a specific cancer syndrome, or has a compelling family history of the disease. As in the prior examples, this is because individual cancer gene testing is expensive and low throughput. One can ask the question whether this is the ideal approach or if we could be screening for these disorders from genome or exome data. Again, we applied sequencing analysis for these genes to the ClinSeq cohort because they were not ascertained for that phenotype. In a published study of 572 exomes ( Johnston et al. 2012 ), updated here to include 850 exomes, we have identified 10 patients with seven distinct cancer susceptibility syndrome mutations. These were mostly familial breast and ovarian cancer ( BRCA1 and BRCA2 ), with one patient each with paraganglioma and pheochromocytoma ( SDHC ) and one with Lynch syndrome ( MSH6 ). What is remarkable about these diagnoses is that only about half of them had a convincing personal or family history of the disease, and thus most would have not been offered testing using the current, hypothesis-testing clinical paradigm. These data suggest that screening for these disorders using genome or exome sequencing could markedly improve our ability to identify such families before they develop or die from these diseases—the ideal of predictive genomic medicine.
Despite these optimistic scenarios and examples, it remains true that our ability to perform true predictive medicine is limited. These limitations include technical factors such as incomplete sequence coverage, imperfect sequence quality, inadequate knowledge regarding the penetrance and expressivity of most variants, uncertain medical approaches and utility of pursuing variants from genomic sequencing, and the poor preparation of most clinicians for addressing genomic concerns in the clinic ( Biesecker 2013 ). Recognizing all of these limitations, it is clear that we are not prepared to launch broad-scale implementation of predictive genomic medicine, nor should all research be structured using the hypothesis-generating approach.
Hypothesis-testing approaches to science and medicine have served us well and should continue. However, the advent of massively parallel sequencing and other high-throughput technologies provides opportunities to undertake hypothesis-generating approaches to science and medicine, which in turn provide unprecedented opportunities for discovery in the research realm. This can allow the discovery of results that were not anticipated or intended by the research design, yet provide critical insights into biology and pathophysiology. Similarly, hypothesis-generating clinical research has the potential to provide these same insights and, in addition, has the potential to provide us with data that will illuminate the full spectrum of genotype–phenotype correlations, eliminating the biases that have limited this understanding in the past. Finally, applying these principles to clinical medicine can provide new pathways to diagnosis and provide the theoretical basis for predictive medicine that can detect disease susceptibility and allow health to be maintained, instead of solely focusing on the treatment of evident disease.
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.157826.113 .
The chief complaint is a brief description of the problem that led the patient to the clinician, such as “I have a cough and fever.”
- Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, et al. 2011. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med 3: 65ra64. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Biesecker LG 2013. Incidental findings are critical for genomics. Am J Hum Genet 92: 648–651 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Biesecker LG, Mullikin JC, Facio FM, Turner C, Cherukuri PF, Blakesley RW, Bouffard GG, Chines PS, Cruz P, Hansen NF, et al. 2009. The ClinSeq Project: Piloting large-scale genome sequencing for research in genomic medicine. Genome Res 19: 1665–1674 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Hennekam RC, Biesecker LG 2012. Next-generation sequencing demands next-generation phenotyping. Hum Mutat 33: 884–886 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [ DOI ] [ PubMed ] [ Google Scholar ]
- Johnston JJ, Rubinstein WS, Facio FM, Ng D, Singh LN, Teer JK, Mullikin JC, Biesecker LG 2012. Secondary variants in individuals undergoing exome sequencing: Screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet 91: 97–108 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Mardis ER 2008. The impact of next-generation sequencing technology on genetics. Trends Genet 24: 133–141 [ DOI ] [ PubMed ] [ Google Scholar ]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, et al. 2010. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 86: 749–764 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [ DOI ] [ PubMed ] [ Google Scholar ]
- Sloan JL, Johnston JJ, Manoli I, Chandler RJ, Krause C, Carrillo-Carrasco N, Chandrasekaran SD, Sysol JR, O'Brien K, Hauser NS, et al. 2011. Exome sequencing identifies ACSF3 as a cause of combined malonic and methylmalonic aciduria. Nat Genet 43: 883–886 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- View on publisher site
- PDF (444.0 KB)
- Collections
Similar articles
Cited by other articles, links to ncbi databases.
- Download .nbib .nbib
- Format: AMA APA MLA NLM
Add to Collections

IMAGES
VIDEO