
Research Methodology
- Introduction to Research Methodology
- Research Approaches
- Concepts of Theory and Empiricism
- Characteristics of scientific method
- Understanding the Language of Research
- 11 Steps in Research Process
- Research Design
- Different Research Designs
- Compare and Contrast the Main Types of Research Designs
- Cross-sectional research design
- Qualitative and Quantitative Research
- Descriptive Research VS Qualitative Research
- Experimental Research VS Quantitative Research
- Sampling Design
- Probability VS Non-Probability Sampling
40 MCQ on Research Methodology
- MCQ on research Process
- MCQ on Research Design
- 18 MCQ on Quantitative Research
- 30 MCQ on Qualitative Research
- 45 MCQ on Sampling Methods
- 20 MCQ on Principles And Planning For Research
Q1. Which of the following statement is correct? (A) Reliability ensures the validity (B) Validity ensures reliability (C) Reliability and validity are independent of each other (D) Reliability does not depend on objectivity
Answer: (C)
Q2. Which of the following statements is correct? (A) Objectives of research are stated in first chapter of the thesis (B) Researcher must possess analytical ability (C) Variability is the source of problem (D) All the above
Answer: (D)
Q3. The first step of research is: (A) Selecting a problem (B) Searching a problem (C) Finding a problem (D) Identifying a problem
Q4. Research can be conducted by a person who: (A) holds a postgraduate degree (B) has studied research methodology (C) possesses thinking and reasoning ability (D) is a hard worker
Answer: (B)
Q5. Research can be classified as: (A) Basic, Applied and Action Research (B) Philosophical, Historical, Survey and Experimental Research (C) Quantitative and Qualitative Research (D) All the above
Q6. To test null hypothesis, a researcher uses: (A) t test (B) ANOVA (C) X 2 (D) factorial analysis
Answer: (B)
Q7. Bibliography given in a research report: (A) shows vast knowledge of the researcher (B) helps those interested in further research (C) has no relevance to research (D) all the above
Q8. A research problem is feasible only when: (A) it has utility and relevance (B) it is researchable (C) it is new and adds something to knowledge (D) all the above
Q9. The study in which the investigators attempt to trace an effect is known as: (A) Survey Research (B) Summative Research (C) Historical Research (D) ‘Ex-post Facto’ Research
Answer: (D)
Q10. Generalized conclusion on the basis of a sample is technically known as: (A) Data analysis and interpretation (B) Parameter inference (C) Statistical inference (D) All of the above
Answer: (A)
Q11. Fundamental research reflects the ability to: (A) Synthesize new ideals (B) Expound new principles (C) Evaluate the existing material concerning research (D) Study the existing literature regarding various topics
Q12. The main characteristic of scientific research is: (A) empirical (B) theoretical (C) experimental (D) all of the above
Q13. Authenticity of a research finding is its: (A) Originality (B) Validity (C) Objectivity (D) All of the above
Q14. Which technique is generally followed when the population is finite? (A) Area Sampling Technique (B) Purposive Sampling Technique (C) Systematic Sampling Technique (D) None of the above
Q15. Research problem is selected from the stand point of: (A) Researcher’s interest (B) Financial support (C) Social relevance (D) Availability of relevant literature
Q16. The research is always – (A) verifying the old knowledge (B) exploring new knowledge (C) filling the gap between knowledge (D) all of these
Q17. Research is (A) Searching again and again (B) Finding a solution to any problem (C) Working in a scientific way to search for the truth of any problem (D) None of the above
Q20. A common test in research demands much priority on (A) Reliability (B) Useability (C) Objectivity (D) All of the above
Q21. Which of the following is the first step in starting the research process? (A) Searching sources of information to locate the problem. (B) Survey of related literature (C) Identification of the problem (D) Searching for solutions to the problem
Answer: (C)
Q22. Which correlation coefficient best explains the relationship between creativity and intelligence? (A) 1.00 (B) 0.6 (C) 0.5 (D) 0.3
Q23. Manipulation is always a part of (A) Historical research (B) Fundamental research (C) Descriptive research (D) Experimental research
Explanation: In experimental research, researchers deliberately manipulate one or more independent variables to observe their effects on dependent variables. The goal is to establish cause-and-effect relationships and test hypotheses. This type of research often involves control groups and random assignment to ensure the validity of the findings. Manipulation is an essential aspect of experimental research to assess the impact of specific variables and draw conclusions about their influence on the outcome.
Q24. The research which is exploring new facts through the study of the past is called (A) Philosophical research (B) Historical research (C) Mythological research (D) Content analysis
Q25. A null hypothesis is (A) when there is no difference between the variables (B) the same as research hypothesis (C) subjective in nature (D) when there is difference between the variables
Q26. We use Factorial Analysis: (A) To know the relationship between two variables (B) To test the Hypothesis (C) To know the difference between two variables (D) To know the difference among the many variables
Explanation: Factorial analysis, specifically factorial analysis of variance (ANOVA), is used to investigate the effects of two or more independent variables on a dependent variable. It helps to determine whether there are significant differences or interactions among the independent variables and their combined effects on the dependent variable.
Q27. Which of the following is classified in the category of the developmental research? (A) Philosophical research (B) Action research (C) Descriptive research (D) All the above
Q28. Action-research is: (A) An applied research (B) A research carried out to solve immediate problems (C) A longitudinal research (D) All the above
Explanation: Action research is an approach to research that encompasses all the options mentioned. It is an applied research method where researchers work collaboratively with practitioners or stakeholders to address immediate problems or issues in a real-world context. It is often conducted over a period of time, making it a longitudinal research approach. So, all the options (A) An applied research, (B) A research carried out to solve immediate problems, and (C) A longitudinal research are correct when describing action research.
Q29. The basis on which assumptions are formulated: (A) Cultural background of the country (B) Universities (C) Specific characteristics of the castes (D) All of these
Q30. How can the objectivity of the research be enhanced? (A) Through its impartiality (B) Through its reliability (C) Through its validity (D) All of these
Q31. A research problem is not feasible only when: (A) it is researchable (B) it is new and adds something to the knowledge (C) it consists of independent and dependent var i ables (D) it has utility and relevance
Explanation: A research problem is considered feasible when it can be studied and investigated using appropriate research methods and resources. The presence of independent and dependent variables is not a factor that determines the feasibility of a research problem. Instead, it is an essential component of a well-defined research problem that helps in formulating research questions or hypotheses. Feasibility depends on whether the research problem can be addressed and answered within the constraints of available time, resources, and methods. Options (A), (B), and (D) are more relevant to the feasibility of a research problem.
Q32. The process not needed in experimental research is: (A) Observation (B) Manipulation and replication (C) Controlling (D) Reference collection
In experimental research, reference collection is not a part of the process.
Q33. When a research problem is related to heterogeneous population, the most suitable sampling method is: (A) Cluster Sampling (B) Stratified Sampling (C) Convenient Sampling (D) Lottery Method
Explanation: When a research problem involves a heterogeneous population, stratified sampling is the most suitable sampling method. Stratified sampling involves dividing the population into subgroups or strata based on certain characteristics or variables. Each stratum represents a relatively homogeneous subset of the population. Then, a random sample is taken from each stratum in proportion to its size or importance in the population. This method ensures that the sample is representative of the diversity present in the population and allows for more precise estimates of population parameters for each subgroup.
Q34. Generalised conclusion on the basis of a sample is technically known as: (A) Data analysis and interpretation (B) Parameter inference (C) Statistical inference (D) All of the above
Explanation: Generalized conclusions based on a sample are achieved through statistical inference. It involves using sample data to make inferences or predictions about a larger population. Statistical inference helps researchers draw conclusions, estimate parameters, and test hypotheses about the population from which the sample was taken. It is a fundamental concept in statistics and plays a crucial role in various fields, including research, data analysis, and decision-making.
Q35. The experimental study is based on
(A) The manipulation of variables (B) Conceptual parameters (C) Replication of research (D) Survey of literature
Q36. Which one is called non-probability sampling? (A) Cluster sampling (B) Quota sampling (C) Systematic sampling (D) Stratified random sampling
Q37. Formulation of hypothesis may NOT be required in: (A) Survey method (B) Historical studies (C) Experimental studies (D) Normative studies
Q38. Field-work-based research is classified as: (A) Empirical (B) Historical (C) Experimental (D) Biographical
Q39. Which of the following sampling method is appropriate to study the prevalence of AIDS amongst male and female in India in 1976, 1986, 1996 and 2006? (A) Cluster sampling (B) Systematic sampling (C) Quota sampling (D) Stratified random sampling
Q40. The research that applies the laws at the time of field study to draw more and more clear ideas about the problem is: (A) Applied research (B) Action research (C) Experimental research (D) None of these
Answer: (A)
Probability and Statistics Questions and Answers – Testing of Hypothesis
This set of Probability and Statistics Multiple Choice Questions & Answers (MCQs) focuses on “Testing of Hypothesis”.
Sanfoundry Global Education & Learning Series – Probability and Statistics.
To practice all areas of Probability and Statistics, here is complete set of 1000+ Multiple Choice Questions and Answers .
- Check Probability and Statistics Books
- Practice Numerical Methods MCQ
- Practice Engineering Mathematics MCQ
- Apply for 1st Year Engineering Internship
Recommended Articles:
- Probability and Statistics Questions and Answers – Testing of Hypothesis Concerning Single Population Mean
- Probability and Statistics Questions and Answers – Probability Distributions – 1
- Probability and Statistics Questions and Answers – Sampling Distribution of Means
- Probability and Statistics Questions and Answers – Sampling Distribution – 2
- Probability and Statistics Questions and Answers – Chi-Squared Distribution
- Probability and Statistics Questions and Answers – Sampling Distribution – 1
- Probability and Statistics MCQ (Multiple Choice Questions)
- Probability and Statistics Questions and Answers – Sampling Distribution of Proportions
- Probability and Statistics Questions and Answers – F-Distribution
- Probability and Statistics Questions and Answers – Probability Distributions – 2
- Probability and Statistics MCQ Questions
- Mechanical Behaviour & Testing of Materials MCQ Questions
- Probability MCQ Questions
- JavaScript MCQ Questions
- Statistical Quality Control MCQ Questions
- Visual Basic MCQ Questions
- C# Programs on Functions
- String Programs in C++
- Linked List Programs in C
- JUnit MCQ Questions

Research Methodology Quiz | MCQ (Multiple Choice Questions)
In order to enhance your understanding of research methodology, we have made thought-provoking quiz featuring multiple-choice questions.
The quiz aimed to sharpen your critical thinking skills and reinforce our grasp on essential concepts in the realm of research. By actively participating in this exercise, we deepened your appreciation for the significance of selecting the right research methods to achieve reliable and meaningful results.
Other articles
Related posts, alternative hypothesis: types and examples, causal research: examples, benefits, and practical tips, 7 types of observational studies | examples, writing an introduction for a research paper: a guide (with examples), clinical research design: elements and importance, dependent variable in research: examples, what is an independent variable, how to write a conclusion for research paper | examples, six useful tips for finding research gap, example of abstract for your research paper: tips, dos, and don’ts, leave a reply cancel reply.
Research Methods- multiple choice exam questions

Students also viewed


- Definitions
Verbal Ability
- Interview Q
| 1) Who was the author of the book named "Methods in Social Research"? c) Goode and HaltThe book named "Methods in Social Research" was authored by Goode and Hatt on Dec 01, 1952, which was specifically aimed to improve student's knowledge as well as response skills. a) Association among variablesMainly the correlational analysis focus on finding the association between one or more quantitative independent variables and one or more quantitative dependent variables. d) Research designA conceptual framework can be understood as a Research design that you require before research. d) To help an applicant in becoming a renowned educationalistEducational research can be defined as an assurance for reviewing and improving educational practice, which will result in becoming a renowned educationalist. c) Collecting data with bottom-up empirical evidence.In qualitative research, we use an inductive methodology that starts from particular to general. In other words, we study society from the bottom, then move upward to make the theories. d) All of the aboveIn random sampling, for each element of the set, there exist a possibility to get selected. c) Ex-post facto methodMainly in the ex-post facto method, the existing groups with qualities are compared on some dependent variable. It is also known as quasi-experimental for the fact that instead of randomly assigning the subjects, they are grouped on the basis of a particular characteristic or trait. d) All of the aboveTippit table was first published by L.H.C Tippett in 1927. b) Formulating a research questionBefore starting with research, it is necessary to have a research question or a topic because once the problem is identified, then we can decide the research design. c) A research dissertationThe format of thesis writing is similar to that of a research dissertation, or we can simply say that dissertation is another word for a thesis. d) Its sole purpose is the production of knowledgeParticipatory action research is a kind of research that stresses participation and action. b) It is only the null hypothesis that can be tested.Hypotheses testing evaluates its plausibility by using sample data. b) The null hypotheses get rejected even if it is trueThe Type-I Error can be defined as the first kind of error. d) All of the above.No explanation. a) Long-term researchIn general, the longitudinal approach is long-term research in which the researchers keep on examining similar individuals to detect if any change has occurred over a while. b) Following an aimNo explanation. a) How well are we doing?Instead of focusing on the process, the evaluation research measures the consequences of the process, for example, if the objectives are met or not. d) Research is not a processResearch is an inspired and systematic work that is undertaken by the researchers to intensify expertise. d) All of the aboveResearch is an inspired and systematic work that is undertaken by the researchers to intensify expertise. b) To bring out the holistic approach to researchParticularly in interdisciplinary research, it combines two or more hypothetical disciplines into one activity. d) Eliminate spurious relationsScientific research aims to build knowledge by hypothesizing new theories and discovering laws. c) QuestionnaireSince it is an urban area, so there is a probability of literacy amongst a greater number of people. Also, there would be numerous questions over the ruling period of a political party, which cannot be simply answered by rating. The rating can only be considered if any political party has done some work, which is why the Questionnaire is used. b) Historical ResearchOne cannot generalize historical research in the USA, which has been done in India. c) By research objectivesResearch objectives concisely demonstrate what we are trying to achieve through the research. c) Has studied research methodologyAnyone who has studied the research methodology can undergo the research. c) ObservationMainly the research method comprises strategies, processes or techniques that are being utilized to collect the data or evidence so as to reveal new information or create a better understanding of a topic. d) All of the aboveA research problem can be defined as a statement about the area of interest, a condition that is required to be improved, a difficulty that has to be eradicated, or any disquieting question existing in scholarly literature, in theory, or in practice that points to be solved. d) How are various parts related to the whole?A circle graph helps in visualizing information as well as the data. b) ObjectivityNo explanation. a) Quota samplingIn non-probability sampling, all the members do not get an equal opportunity to participate in the study. a) Reducing punctuations as well as grammatical errors to minimalist Select the answers from the codes given below: B. a), b), c) and d)All of the above. a) Research refers to a series of systematic activity or activities undertaken to find out the solution to a problem. Select the correct answer from the codes given below: A. a), b), c) and d)All of the above. b) Fundamental ResearchJean Piaget, in his cognitive-developmental theory, proposed the idea that children can actively construct knowledge simply by exploring and manipulating the world around them. d) Introduction; Literature Review; Research Methodology; Results; Discussions and ConclusionsThe core elements of the dissertation are as follows: Introduction; Literature Review; Research Methodology; Results; Discussions and Conclusions d) A sampling of people, newspapers, television programs etc.In general, sampling in case study research involves decisions made by the researchers regarding the strategies of sampling, the number of case studies, and the definition of the unit of analysis. a) Systematic Sampling TechniqueSystematic sampling can be understood as a probability sampling method in which the members of the population are selected by the researchers at a regular interval. a) Social relevanceNo explanation. c) Can be one-tailed as well as two-tailed depending on the hypothesesAn F-test corresponds to a statistical test in which the test statistic has an F-distribution under the null hypothesis. a) CensusCensus is an official survey that keeps track of the population data. b) ObservationNo explanation. d) It contains dependent and independent variablesA research problem can be defined as a statement about the concerned area, a condition needed to be improved, a difficulty that has to be eliminated, or a troubling question existing in scholarly literature, in theory, or in practice pointing towards the need of delivering a deliberate investigation. d) All of the aboveThe research objectives must be concisely described before starting the research as it illustrates what we are going to achieve as an end result after the accomplishment. c) A kind of research being carried out to solve a specific problemIn general, action research is termed as a philosophy or a research methodology, which is implemented in social sciences. a) The cultural background of the countryAn assumption can be identified as an unexamined belief, which we contemplate without even comprehending it. Also, the conclusions that we draw are often based on assumptions. d) All of the aboveNo explanation. b) To understand the difference between two variablesFactor analysis can be understood as a statistical method that defines the variability between two variables in terms of factors, which are nothing but unobserved variables. a) ManipulationIn an experimental research design, whenever the independent variables (i.e., treatment variables or factors) decisively get altered by researchers, then that process is termed as an experimental manipulation. d) Professional AttitudeA professional attitude is an ability that inclines you to manage your time, portray a leadership quality, make you self-determined and persistent. b) Human RelationsThe term sociogram can be defined as a graphical representation of human relation that portrays the social links formed by one particular person. c) Objective ObservationThe research process comprises classifying, locating, evaluating, and investigating the data, which is required to support your research question, followed by developing and expressing your ideas. |

- Send your Feedback to [email protected]
Help Others, Please Share

Learn Latest Tutorials
Transact-SQL
Reinforcement Learning
R Programming
React Native
Python Design Patterns
Python Pillow
Python Turtle
Preparation

Interview Questions

Company Questions
Trending Technologies
Artificial Intelligence
Cloud Computing
Data Science
Machine Learning
B.Tech / MCA
Data Structures
Operating System
Computer Network
Compiler Design
Computer Organization
Discrete Mathematics
Ethical Hacking
Computer Graphics
Software Engineering
Web Technology
Cyber Security
C Programming
Control System
Data Mining
Data Warehouse


In order to continue enjoying our site, we ask that you confirm your identity as a human. Thank you very much for your cooperation.
University Courses
Introduction to Psychology Practice Tests
Introduction to Psychology Online Tests
Research Hypothesis Multiple Choice Questions (MCQ) PDF Download
The Research Hypothesis Multiple Choice Questions (MCQ Quiz) with Answers PDF , Research Hypothesis MCQ PDF e-Book download to practice Introduction to Psychology Tests . Study Psychological Science Multiple Choice Questions and Answers (MCQs) , Research Hypothesis quiz answers PDF to study MSc in psychology courses. The Research Hypothesis MCQ App Download: Free learning app for psychological science, scientific method, ensuring that research is ethical test prep for online college courses.
The MCQ: A attribute, presuming different values among different people in different times or places, known as; "Research Hypothesis" App Download (Free) with answers: Symbol; Attribute; Scientific; Variable; to study MSc in psychology courses. Practice Research Hypothesis Quiz Questions , download Apple e-Book (Free Sample) for accelerated bachelors degree online.
Research Hypothesis MCQs: Questions and Answers PDF Download
A theoretical ideas that form the basis of research hypothesis is:
- Research hypothesis
- Research analysis
- Conceptual variables
- Composed data
A attribute, presuming different values among different people in different times or places, known as:
The concept which form the basis of a research hypothesis are known as:
- Research Method
- Theory of organisms
A concrete statement, prediction of what may happen in a study, termed as:
- Research Tools
A variables that consisting of a numbers that represent the conceptual variables are known as:
- Measured variables
- Non vulnerable variables
Introduction To Psychology Practice Tests
Research hypothesis learning app: free download android & ios.
The App: Research Hypothesis MCQs App to study Research Hypothesis textbook, Introduction to Psychology MCQ App, and RF Electronics MCQ App. The "Research Hypothesis MCQs" App to free download iOS & Android Apps includes complete analytics with interactive assessments. Download App Store & Play Store learning Apps & enjoy 100% functionality with subscriptions!
Research Hypothesis App (Android & iOS)
Introduction to Psychology App (iOS & Android)
RF Electronics App (Android & iOS)
Educational Psychology App (iOS & Android)
Histology MCQs eBook Download

Histology MCQ Book PDF
Microbiology Practice Questions
- Basic Mycology MCQs
- Classification of Medically important Bacteria MCQs
- Classification of Viruses MCQs
- Clinical Virology MCQs
- Drugs and Vaccines MCQs
- Genetics of Bacterial Cells MCQs
- Genetics of Viruses MCQs
- Growth of Bacterial Cells MCQs
- Brains, Bodies, and Behavior Quiz
- Emotions and Motivations Quiz
- Growing and Developing Quiz
- Introduction to Psychology Quiz
- Learning Phychology Quiz
- Personality Quiz
- Psychological Science Quiz
- Remembering and Judging Quiz
Microbiology MCQ Questions
- Inflammation of cornea usually occurs in wearing contact lenses resulting inflammation is known as
- What is estimated diameter of Caliciviruses?
- Absence of skin and other organs is a disease caused by
- Anthrax is caused by gram-positive rod named as
- Shigellosis is a disease found particularly in
Research Hypothesis MCQs Book Questions
- Two areas of 'hypothalamus' are known as:
- Personality is a derivation of our;
- Most important communicator of emotion is:
- Hormonal surge in developmental stage is related with;
- Light enters in our eye through;

- Request new password
- Create a new account
Research Methodology
Student resources, multiple choice questions.
Research: A Way of Thinking
The Research Process: A Quick Glance
Reviewing the Literature
Formulating a Research Problem
Identifying Variables
Constructing Hypotheses
The Research Design
Selecting a Study Design
Selecting a Method of Data Collection
Collecting Data Using Attitudinal Scales
Establishing the Validity and Reliability of a Research Instrument
Selecting a Sample
Writing a Research Proposal
Considering Ethical Issues in Data Collection
Processing Data
Displaying Data
Writing a Research Report
430+ Research Methodology (RM) Solved MCQs
| 1. | |
| A. | Wilkinson |
| B. | CR Kothari |
| C. | Kerlinger |
| D. | Goode and Halt |
| Answer» D. Goode and Halt | |
| 2. | |
| A. | Marshall |
| B. | P.V. Young |
| C. | Emory |
| D. | Kerlinger |
| Answer» C. Emory | |
| 3. | |
| A. | Young |
| B. | Kerlinger |
| C. | Kothari |
| D. | Emory |
| Answer» A. Young | |
| 4. | |
| A. | Experiment |
| B. | Observation |
| C. | Deduction |
| D. | Scientific method |
| Answer» D. Scientific method | |
| 5. | |
| A. | Deduction |
| B. | Scientific method |
| C. | Observation |
| D. | experience |
| Answer» B. Scientific method | |
| 6. | |
| A. | Objectivity |
| B. | Ethics |
| C. | Proposition |
| D. | Neutrality |
| Answer» A. Objectivity | |
| 7. | |
| A. | Induction |
| B. | Deduction |
| C. | Research |
| D. | Experiment |
| Answer» A. Induction | |
| 8. | |
| A. | Belief |
| B. | Value |
| C. | Objectivity |
| D. | Subjectivity |
| Answer» C. Objectivity | |
| 9. | |
| A. | Induction |
| B. | deduction |
| C. | Observation |
| D. | experience |
| Answer» B. deduction | |
| 10. | |
| A. | Caroline |
| B. | P.V.Young |
| C. | Dewey John |
| D. | Emory |
| Answer» B. P.V.Young | |
| 11. | |
| A. | Facts |
| B. | Values |
| C. | Theory |
| D. | Generalization |
| Answer» C. Theory | |
| 12. | |
| A. | Jack Gibbs |
| B. | PV Young |
| C. | Black |
| D. | Rose Arnold |
| Answer» B. PV Young | |
| 13. | |
| A. | Black James and Champion |
| B. | P.V. Young |
| C. | Emory |
| D. | Gibbes |
| Answer» A. Black James and Champion | |
| 14. | |
| A. | Theory |
| B. | Value |
| C. | Fact |
| D. | Statement |
| Answer» C. Fact | |
| 15. | |
| A. | Good and Hatt |
| B. | Emory |
| C. | P.V. Young |
| D. | Claver |
| Answer» A. Good and Hatt | |
| 16. | |
| A. | Concept |
| B. | Variable |
| C. | Model |
| D. | Facts |
| Answer» C. Model | |
| 17. | |
| A. | Objects |
| B. | Human beings |
| C. | Living things |
| D. | Non living things |
| Answer» B. Human beings | |
| 18. | |
| A. | Natural and Social |
| B. | Natural and Physical |
| C. | Physical and Mental |
| D. | Social and Physical |
| Answer» A. Natural and Social | |
| 19. | |
| A. | Causal Connection |
| B. | reason |
| C. | Interaction |
| D. | Objectives |
| Answer» A. Causal Connection | |
| 20. | |
| A. | Explain |
| B. | diagnosis |
| C. | Recommend |
| D. | Formulate |
| Answer» B. diagnosis | |
| 21. | |
| A. | Integration |
| B. | Social Harmony |
| C. | National Integration |
| D. | Social Equality |
| Answer» A. Integration | |
| 22. | |
| A. | Unit |
| B. | design |
| C. | Random |
| D. | Census |
| Answer» B. design | |
| 23. | |
| A. | Objectivity |
| B. | Specificity |
| C. | Values |
| D. | Facts |
| Answer» A. Objectivity | |
| 24. | |
| A. | Purpose |
| B. | Intent |
| C. | Methodology |
| D. | Techniques |
| Answer» B. Intent | |
| 25. | |
| A. | Pure Research |
| B. | Action Research |
| C. | Pilot study |
| D. | Survey |
| Answer» A. Pure Research | |
| 26. | |
| A. | Pure Research |
| B. | Survey |
| C. | Action Research |
| D. | Long term Research |
| Answer» B. Survey | |
| 27. | |
| A. | Survey |
| B. | Action research |
| C. | Analytical research |
| D. | Pilot study |
| Answer» C. Analytical research | |
| 28. | |
| A. | Fundamental Research |
| B. | Analytical Research |
| C. | Survey |
| D. | Action Research |
| Answer» D. Action Research | |
| 29. | |
| A. | Action Research |
| B. | Survey |
| C. | Pilot study |
| D. | Pure Research |
| Answer» D. Pure Research | |
| 30. | |
| A. | Quantitative |
| B. | Qualitative |
| C. | Pure |
| D. | applied |
| Answer» B. Qualitative | |
| 31. | |
| A. | Empirical research |
| B. | Conceptual Research |
| C. | Quantitative research |
| D. | Qualitative research |
| Answer» B. Conceptual Research | |
| 32. | |
| A. | Clinical or diagnostic |
| B. | Causal |
| C. | Analytical |
| D. | Qualitative |
| Answer» A. Clinical or diagnostic | |
| 33. | |
| A. | Field study |
| B. | Survey |
| C. | Laboratory Research |
| D. | Empirical Research |
| Answer» C. Laboratory Research | |
| 34. | |
| A. | Clinical Research |
| B. | Experimental Research |
| C. | Laboratory Research |
| D. | Empirical Research |
| Answer» D. Empirical Research | |
| 35. | |
| A. | Survey |
| B. | Empirical |
| C. | Clinical |
| D. | Diagnostic |
| Answer» A. Survey | |
| 36. | |
| A. | Ostle |
| B. | Richard |
| C. | Karl Pearson |
| D. | Kerlinger |
| Answer» C. Karl Pearson | |
| 37. | |
| A. | Redmen and Mory |
| B. | P.V.Young |
| C. | Robert C meir |
| D. | Harold Dazier |
| Answer» A. Redmen and Mory | |
| 38. | |
| A. | Technique |
| B. | Operations |
| C. | Research methodology |
| D. | Research Process |
| Answer» C. Research methodology | |
| 39. | |
| A. | Slow |
| B. | Fast |
| C. | Narrow |
| D. | Systematic |
| Answer» D. Systematic | |
| 40. | |
| A. | Logical |
| B. | Non logical |
| C. | Narrow |
| D. | Systematic |
| Answer» A. Logical | |
| 41. | |
| A. | Delta Kappan |
| B. | James Harold Fox |
| C. | P.V.Young |
| D. | Karl Popper |
| Answer» B. James Harold Fox | |
| 42. | |
| A. | Problem |
| B. | Experiment |
| C. | Research Techniques |
| D. | Research methodology |
| Answer» D. Research methodology | |
| 43. | |
| A. | Field Study |
| B. | diagnosis tic study |
| C. | Action study |
| D. | Pilot study |
| Answer» B. diagnosis tic study | |
| 44. | |
| A. | Social Science Research |
| B. | Experience Survey |
| C. | Problem formulation |
| D. | diagnostic study |
| Answer» A. Social Science Research | |
| 45. | |
| A. | P.V. Young |
| B. | Kerlinger |
| C. | Emory |
| D. | Clover Vernon |
| Answer» B. Kerlinger | |
| 46. | |
| A. | Black James and Champions |
| B. | P.V. Young |
| C. | Mortan Kaplan |
| D. | William Emory |
| Answer» A. Black James and Champions | |
| 47. | |
| A. | Best John |
| B. | Emory |
| C. | Clover |
| D. | P.V. Young |
| Answer» D. P.V. Young | |
| 48. | |
| A. | Belief |
| B. | Value |
| C. | Confidence |
| D. | Overconfidence |
| Answer» D. Overconfidence | |
| 49. | |
| A. | Velocity |
| B. | Momentum |
| C. | Frequency |
| D. | gravity |
| Answer» C. Frequency | |
| 50. | |
| A. | Research degree |
| B. | Research Academy |
| C. | Research Labs |
| D. | Research Problems |
| Answer» A. Research degree | |
| 51. | |
| A. | Book |
| B. | Journal |
| C. | News Paper |
| D. | Census Report |
| Answer» C. News Paper | |
| 52. | |
| A. | Lack of sufficient number of Universities |
| B. | Lack of sufficient research guides |
| C. | Lack of sufficient Fund |
| D. | Lack of scientific training in research |
| Answer» D. Lack of scientific training in research | |
| 53. | |
| A. | Indian Council for Survey and Research |
| B. | Indian Council for strategic Research |
| C. | Indian Council for Social Science Research |
| D. | Inter National Council for Social Science Research |
| Answer» C. Indian Council for Social Science Research | |
| 54. | |
| A. | University Grants Commission |
| B. | Union Government Commission |
| C. | University Governance Council |
| D. | Union government Council |
| Answer» A. University Grants Commission | |
| 55. | |
| A. | Junior Research Functions |
| B. | Junior Research Fellowship |
| C. | Junior Fellowship |
| D. | None of the above |
| Answer» B. Junior Research Fellowship | |
| 56. | |
| A. | Formulation of a problem |
| B. | Collection of Data |
| C. | Editing and Coding |
| D. | Selection of a problem |
| Answer» D. Selection of a problem | |
| 57. | |
| A. | Fully solved |
| B. | Not solved |
| C. | Cannot be solved |
| D. | half- solved |
| Answer» D. half- solved | |
| 58. | |
| A. | Schools and Colleges |
| B. | Class Room Lectures |
| C. | Play grounds |
| D. | Infra structures |
| Answer» B. Class Room Lectures | |
| 59. | |
| A. | Observation |
| B. | Problem |
| C. | Data |
| D. | Experiment |
| Answer» B. Problem | |
| 60. | |
| A. | Solution |
| B. | Examination |
| C. | Problem formulation |
| D. | Problem Solving |
| Answer» C. Problem formulation | |
| 61. | |
| A. | Very Common |
| B. | Overdone |
| C. | Easy one |
| D. | rare |
| Answer» B. Overdone | |
| 62. | |
| A. | Statement of the problem |
| B. | Gathering of Data |
| C. | Measurement |
| D. | Survey |
| Answer» A. Statement of the problem | |
| 63. | |
| A. | Professor |
| B. | Tutor |
| C. | HOD |
| D. | Guide |
| Answer» D. Guide | |
| 64. | |
| A. | Statement of the problem |
| B. | Understanding the nature of the problem |
| C. | Survey |
| D. | Discussions |
| Answer» B. Understanding the nature of the problem | |
| 65. | |
| A. | Statement of the problem |
| B. | Understanding the nature of the problem |
| C. | Survey the available literature |
| D. | Discussion |
| Answer» C. Survey the available literature | |
| 66. | |
| A. | Survey |
| B. | Discussion |
| C. | Literature survey |
| D. | Re Phrasing the Research problem |
| Answer» D. Re Phrasing the Research problem | |
| 67. | |
| A. | Title |
| B. | Index |
| C. | Bibliography |
| D. | Concepts |
| Answer» A. Title | |
| 68. | |
| A. | Questions to be answered |
| B. | methods |
| C. | Techniques |
| D. | methodology |
| Answer» A. Questions to be answered | |
| 69. | |
| A. | Speed |
| B. | Facts |
| C. | Values |
| D. | Novelty |
| Answer» D. Novelty | |
| 70. | |
| A. | Originality |
| B. | Values |
| C. | Coherence |
| D. | Facts |
| Answer» A. Originality | |
| 71. | |
| A. | Academic and Non academic |
| B. | Cultivation |
| C. | Academic |
| D. | Utilitarian |
| Answer» B. Cultivation | |
| 72. | |
| A. | Information |
| B. | firsthand knowledge |
| C. | Knowledge and information |
| D. | models |
| Answer» C. Knowledge and information | |
| 73. | |
| A. | Alienation |
| B. | Cohesion |
| C. | mobility |
| D. | Integration |
| Answer» B. Cohesion | |
| 74. | |
| A. | Scientific temper |
| B. | Age |
| C. | Money |
| D. | time |
| Answer» A. Scientific temper | |
| 75. | |
| A. | Secular |
| B. | Totalitarian |
| C. | democratic |
| D. | welfare |
| Answer» D. welfare | |
| 76. | |
| A. | Hypothesis |
| B. | Variable |
| C. | Concept |
| D. | facts |
| Answer» C. Concept | |
| 77. | |
| A. | Abstract and Coherent |
| B. | Concrete and Coherent |
| C. | Abstract and concrete |
| D. | None of the above |
| Answer» C. Abstract and concrete | |
| 78. | |
| A. | 4 |
| B. | 6 |
| C. | 10 |
| D. | 2 |
| Answer» D. 2 | |
| 79. | |
| A. | Observation |
| B. | formulation |
| C. | Theory |
| D. | Postulation |
| Answer» D. Postulation | |
| 80. | |
| A. | Formulation |
| B. | Postulation |
| C. | Intuition |
| D. | Observation |
| Answer» C. Intuition | |
| 81. | |
| A. | guide |
| B. | tools |
| C. | methods |
| D. | Variables |
| Answer» B. tools | |
| 82. | |
| A. | Metaphor |
| B. | Simile |
| C. | Symbols |
| D. | Models |
| Answer» C. Symbols | |
| 83. | |
| A. | Formulation |
| B. | Calculation |
| C. | Abstraction |
| D. | Specification |
| Answer» C. Abstraction | |
| 84. | |
| A. | Verbal |
| B. | Oral |
| C. | Hypothetical |
| D. | Operational |
| Answer» C. Hypothetical | |
| 85. | |
| A. | Kerlinger |
| B. | P.V. Young |
| C. | Aurthur |
| D. | Kaplan |
| Answer» B. P.V. Young | |
| 86. | |
| A. | Same and different |
| B. | Same |
| C. | different |
| D. | None of the above |
| Answer» C. different | |
| 87. | |
| A. | Greek |
| B. | English |
| C. | Latin |
| D. | Many languages |
| Answer» D. Many languages | |
| 88. | |
| A. | Variable |
| B. | Hypothesis |
| C. | Data |
| D. | Concept |
| Answer» B. Hypothesis | |
| 89. | |
| A. | Data |
| B. | Concept |
| C. | Research |
| D. | Hypothesis |
| Answer» D. Hypothesis | |
| 90. | |
| A. | Lund berg |
| B. | Emory |
| C. | Johnson |
| D. | Good and Hatt |
| Answer» D. Good and Hatt | |
| 91. | |
| A. | Good and Hatt |
| B. | Lund berg |
| C. | Emory |
| D. | Orwell |
| Answer» B. Lund berg | |
| 92. | |
| A. | Descriptive |
| B. | Imaginative |
| C. | Relational |
| D. | Variable |
| Answer» A. Descriptive | |
| 93. | |
| A. | Null Hypothesis |
| B. | Working Hypothesis |
| C. | Relational Hypothesis |
| D. | Descriptive Hypothesis |
| Answer» B. Working Hypothesis | |
| 94. | |
| A. | Relational Hypothesis |
| B. | Situational Hypothesis |
| C. | Null Hypothesis |
| D. | Casual Hypothesis |
| Answer» C. Null Hypothesis | |
| 95. | |
| A. | Abstract |
| B. | Dependent |
| C. | Independent |
| D. | Separate |
| Answer» C. Independent | |
| 96. | |
| A. | Independent |
| B. | Dependent |
| C. | Separate |
| D. | Abstract |
| Answer» B. Dependent | |
| 97. | |
| A. | Causal |
| B. | Relational |
| C. | Descriptive |
| D. | Tentative |
| Answer» B. Relational | |
| 98. | |
| A. | One |
| B. | Many |
| C. | Zero |
| D. | None of these |
| Answer» C. Zero | |
| 99. | |
| A. | Statistical Hypothesis |
| B. | Complex Hypothesis |
| C. | Common sense Hypothesis |
| D. | Analytical Hypothesis |
| Answer» C. Common sense Hypothesis | |
| 100. | |
| A. | Null Hypothesis |
| B. | Casual Hypothesis |
| C. | Barren Hypothesis |
| D. | Analytical Hypothesis |
| Answer» D. Analytical Hypothesis | |
- Biology MCQs
- Biology Notes
- __Biotechnology
- __Microbiology
- __Biochemistry
- _Immunology
- _Biology MCQ
- Practice Tests
- _Exam Questions
- _NEET Biology MCQs
Multiple Choice Questions on Research Methodology
Our website uses cookies to improve your experience. Learn more
Contact form

Research Hypothesis In Psychology: Types, & Examples
Saul Mcleod, PhD
Editor-in-Chief for Simply Psychology
BSc (Hons) Psychology, MRes, PhD, University of Manchester
Saul Mcleod, PhD., is a qualified psychology teacher with over 18 years of experience in further and higher education. He has been published in peer-reviewed journals, including the Journal of Clinical Psychology.
Learn about our Editorial Process
Olivia Guy-Evans, MSc
Associate Editor for Simply Psychology
BSc (Hons) Psychology, MSc Psychology of Education
Olivia Guy-Evans is a writer and associate editor for Simply Psychology. She has previously worked in healthcare and educational sectors.
On This Page:
A research hypothesis, in its plural form “hypotheses,” is a specific, testable prediction about the anticipated results of a study, established at its outset. It is a key component of the scientific method .
Hypotheses connect theory to data and guide the research process towards expanding scientific understanding
Some key points about hypotheses:
- A hypothesis expresses an expected pattern or relationship. It connects the variables under investigation.
- It is stated in clear, precise terms before any data collection or analysis occurs. This makes the hypothesis testable.
- A hypothesis must be falsifiable. It should be possible, even if unlikely in practice, to collect data that disconfirms rather than supports the hypothesis.
- Hypotheses guide research. Scientists design studies to explicitly evaluate hypotheses about how nature works.
- For a hypothesis to be valid, it must be testable against empirical evidence. The evidence can then confirm or disprove the testable predictions.
- Hypotheses are informed by background knowledge and observation, but go beyond what is already known to propose an explanation of how or why something occurs.
Predictions typically arise from a thorough knowledge of the research literature, curiosity about real-world problems or implications, and integrating this to advance theory. They build on existing literature while providing new insight.
Types of Research Hypotheses
Alternative hypothesis.
The research hypothesis is often called the alternative or experimental hypothesis in experimental research.
It typically suggests a potential relationship between two key variables: the independent variable, which the researcher manipulates, and the dependent variable, which is measured based on those changes.
The alternative hypothesis states a relationship exists between the two variables being studied (one variable affects the other).
A hypothesis is a testable statement or prediction about the relationship between two or more variables. It is a key component of the scientific method. Some key points about hypotheses:
- Important hypotheses lead to predictions that can be tested empirically. The evidence can then confirm or disprove the testable predictions.
In summary, a hypothesis is a precise, testable statement of what researchers expect to happen in a study and why. Hypotheses connect theory to data and guide the research process towards expanding scientific understanding.
An experimental hypothesis predicts what change(s) will occur in the dependent variable when the independent variable is manipulated.
It states that the results are not due to chance and are significant in supporting the theory being investigated.
The alternative hypothesis can be directional, indicating a specific direction of the effect, or non-directional, suggesting a difference without specifying its nature. It’s what researchers aim to support or demonstrate through their study.
Null Hypothesis
The null hypothesis states no relationship exists between the two variables being studied (one variable does not affect the other). There will be no changes in the dependent variable due to manipulating the independent variable.
It states results are due to chance and are not significant in supporting the idea being investigated.
The null hypothesis, positing no effect or relationship, is a foundational contrast to the research hypothesis in scientific inquiry. It establishes a baseline for statistical testing, promoting objectivity by initiating research from a neutral stance.
Many statistical methods are tailored to test the null hypothesis, determining the likelihood of observed results if no true effect exists.
This dual-hypothesis approach provides clarity, ensuring that research intentions are explicit, and fosters consistency across scientific studies, enhancing the standardization and interpretability of research outcomes.
Nondirectional Hypothesis
A non-directional hypothesis, also known as a two-tailed hypothesis, predicts that there is a difference or relationship between two variables but does not specify the direction of this relationship.
It merely indicates that a change or effect will occur without predicting which group will have higher or lower values.
For example, “There is a difference in performance between Group A and Group B” is a non-directional hypothesis.
Directional Hypothesis
A directional (one-tailed) hypothesis predicts the nature of the effect of the independent variable on the dependent variable. It predicts in which direction the change will take place. (i.e., greater, smaller, less, more)
It specifies whether one variable is greater, lesser, or different from another, rather than just indicating that there’s a difference without specifying its nature.
For example, “Exercise increases weight loss” is a directional hypothesis.

Falsifiability
The Falsification Principle, proposed by Karl Popper , is a way of demarcating science from non-science. It suggests that for a theory or hypothesis to be considered scientific, it must be testable and irrefutable.
Falsifiability emphasizes that scientific claims shouldn’t just be confirmable but should also have the potential to be proven wrong.
It means that there should exist some potential evidence or experiment that could prove the proposition false.
However many confirming instances exist for a theory, it only takes one counter observation to falsify it. For example, the hypothesis that “all swans are white,” can be falsified by observing a black swan.
For Popper, science should attempt to disprove a theory rather than attempt to continually provide evidence to support a research hypothesis.
Can a Hypothesis be Proven?
Hypotheses make probabilistic predictions. They state the expected outcome if a particular relationship exists. However, a study result supporting a hypothesis does not definitively prove it is true.
All studies have limitations. There may be unknown confounding factors or issues that limit the certainty of conclusions. Additional studies may yield different results.
In science, hypotheses can realistically only be supported with some degree of confidence, not proven. The process of science is to incrementally accumulate evidence for and against hypothesized relationships in an ongoing pursuit of better models and explanations that best fit the empirical data. But hypotheses remain open to revision and rejection if that is where the evidence leads.
- Disproving a hypothesis is definitive. Solid disconfirmatory evidence will falsify a hypothesis and require altering or discarding it based on the evidence.
- However, confirming evidence is always open to revision. Other explanations may account for the same results, and additional or contradictory evidence may emerge over time.
We can never 100% prove the alternative hypothesis. Instead, we see if we can disprove, or reject the null hypothesis.
If we reject the null hypothesis, this doesn’t mean that our alternative hypothesis is correct but does support the alternative/experimental hypothesis.
Upon analysis of the results, an alternative hypothesis can be rejected or supported, but it can never be proven to be correct. We must avoid any reference to results proving a theory as this implies 100% certainty, and there is always a chance that evidence may exist which could refute a theory.
How to Write a Hypothesis
- Identify variables . The researcher manipulates the independent variable and the dependent variable is the measured outcome.
- Operationalized the variables being investigated . Operationalization of a hypothesis refers to the process of making the variables physically measurable or testable, e.g. if you are about to study aggression, you might count the number of punches given by participants.
- Decide on a direction for your prediction . If there is evidence in the literature to support a specific effect of the independent variable on the dependent variable, write a directional (one-tailed) hypothesis. If there are limited or ambiguous findings in the literature regarding the effect of the independent variable on the dependent variable, write a non-directional (two-tailed) hypothesis.
- Make it Testable : Ensure your hypothesis can be tested through experimentation or observation. It should be possible to prove it false (principle of falsifiability).
- Clear & concise language . A strong hypothesis is concise (typically one to two sentences long), and formulated using clear and straightforward language, ensuring it’s easily understood and testable.
Consider a hypothesis many teachers might subscribe to: students work better on Monday morning than on Friday afternoon (IV=Day, DV= Standard of work).
Now, if we decide to study this by giving the same group of students a lesson on a Monday morning and a Friday afternoon and then measuring their immediate recall of the material covered in each session, we would end up with the following:
- The alternative hypothesis states that students will recall significantly more information on a Monday morning than on a Friday afternoon.
- The null hypothesis states that there will be no significant difference in the amount recalled on a Monday morning compared to a Friday afternoon. Any difference will be due to chance or confounding factors.
More Examples
- Memory : Participants exposed to classical music during study sessions will recall more items from a list than those who studied in silence.
- Social Psychology : Individuals who frequently engage in social media use will report higher levels of perceived social isolation compared to those who use it infrequently.
- Developmental Psychology : Children who engage in regular imaginative play have better problem-solving skills than those who don’t.
- Clinical Psychology : Cognitive-behavioral therapy will be more effective in reducing symptoms of anxiety over a 6-month period compared to traditional talk therapy.
- Cognitive Psychology : Individuals who multitask between various electronic devices will have shorter attention spans on focused tasks than those who single-task.
- Health Psychology : Patients who practice mindfulness meditation will experience lower levels of chronic pain compared to those who don’t meditate.
- Organizational Psychology : Employees in open-plan offices will report higher levels of stress than those in private offices.
- Behavioral Psychology : Rats rewarded with food after pressing a lever will press it more frequently than rats who receive no reward.
Related Articles
Research Methodology
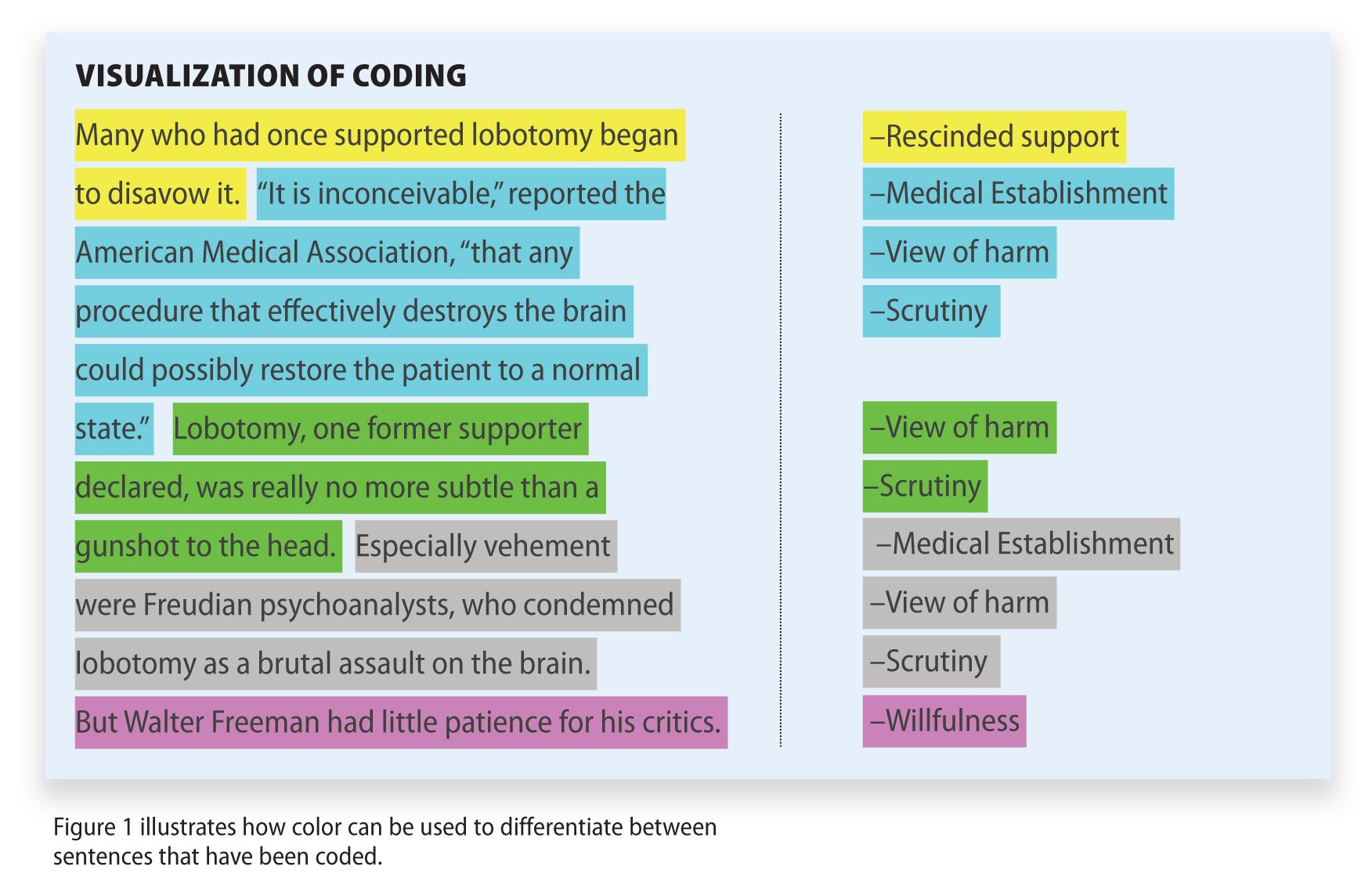
Qualitative Data Coding

What Is a Focus Group?
Cross-Cultural Research Methodology In Psychology

What Is Internal Validity In Research?

Research Methodology , Statistics
What Is Face Validity In Research? Importance & How To Measure

Criterion Validity: Definition & Examples
What is a scientific hypothesis?
It's the initial building block in the scientific method.

Hypothesis basics
What makes a hypothesis testable.
- Types of hypotheses
- Hypothesis versus theory
Additional resources
Bibliography.
A scientific hypothesis is a tentative, testable explanation for a phenomenon in the natural world. It's the initial building block in the scientific method . Many describe it as an "educated guess" based on prior knowledge and observation. While this is true, a hypothesis is more informed than a guess. While an "educated guess" suggests a random prediction based on a person's expertise, developing a hypothesis requires active observation and background research.
The basic idea of a hypothesis is that there is no predetermined outcome. For a solution to be termed a scientific hypothesis, it has to be an idea that can be supported or refuted through carefully crafted experimentation or observation. This concept, called falsifiability and testability, was advanced in the mid-20th century by Austrian-British philosopher Karl Popper in his famous book "The Logic of Scientific Discovery" (Routledge, 1959).
A key function of a hypothesis is to derive predictions about the results of future experiments and then perform those experiments to see whether they support the predictions.
A hypothesis is usually written in the form of an if-then statement, which gives a possibility (if) and explains what may happen because of the possibility (then). The statement could also include "may," according to California State University, Bakersfield .
Here are some examples of hypothesis statements:
- If garlic repels fleas, then a dog that is given garlic every day will not get fleas.
- If sugar causes cavities, then people who eat a lot of candy may be more prone to cavities.
- If ultraviolet light can damage the eyes, then maybe this light can cause blindness.
A useful hypothesis should be testable and falsifiable. That means that it should be possible to prove it wrong. A theory that can't be proved wrong is nonscientific, according to Karl Popper's 1963 book " Conjectures and Refutations ."
An example of an untestable statement is, "Dogs are better than cats." That's because the definition of "better" is vague and subjective. However, an untestable statement can be reworded to make it testable. For example, the previous statement could be changed to this: "Owning a dog is associated with higher levels of physical fitness than owning a cat." With this statement, the researcher can take measures of physical fitness from dog and cat owners and compare the two.
Types of scientific hypotheses

In an experiment, researchers generally state their hypotheses in two ways. The null hypothesis predicts that there will be no relationship between the variables tested, or no difference between the experimental groups. The alternative hypothesis predicts the opposite: that there will be a difference between the experimental groups. This is usually the hypothesis scientists are most interested in, according to the University of Miami .
For example, a null hypothesis might state, "There will be no difference in the rate of muscle growth between people who take a protein supplement and people who don't." The alternative hypothesis would state, "There will be a difference in the rate of muscle growth between people who take a protein supplement and people who don't."
If the results of the experiment show a relationship between the variables, then the null hypothesis has been rejected in favor of the alternative hypothesis, according to the book " Research Methods in Psychology " (BCcampus, 2015).
There are other ways to describe an alternative hypothesis. The alternative hypothesis above does not specify a direction of the effect, only that there will be a difference between the two groups. That type of prediction is called a two-tailed hypothesis. If a hypothesis specifies a certain direction — for example, that people who take a protein supplement will gain more muscle than people who don't — it is called a one-tailed hypothesis, according to William M. K. Trochim , a professor of Policy Analysis and Management at Cornell University.
Sometimes, errors take place during an experiment. These errors can happen in one of two ways. A type I error is when the null hypothesis is rejected when it is true. This is also known as a false positive. A type II error occurs when the null hypothesis is not rejected when it is false. This is also known as a false negative, according to the University of California, Berkeley .
A hypothesis can be rejected or modified, but it can never be proved correct 100% of the time. For example, a scientist can form a hypothesis stating that if a certain type of tomato has a gene for red pigment, that type of tomato will be red. During research, the scientist then finds that each tomato of this type is red. Though the findings confirm the hypothesis, there may be a tomato of that type somewhere in the world that isn't red. Thus, the hypothesis is true, but it may not be true 100% of the time.
Scientific theory vs. scientific hypothesis
The best hypotheses are simple. They deal with a relatively narrow set of phenomena. But theories are broader; they generally combine multiple hypotheses into a general explanation for a wide range of phenomena, according to the University of California, Berkeley . For example, a hypothesis might state, "If animals adapt to suit their environments, then birds that live on islands with lots of seeds to eat will have differently shaped beaks than birds that live on islands with lots of insects to eat." After testing many hypotheses like these, Charles Darwin formulated an overarching theory: the theory of evolution by natural selection.
"Theories are the ways that we make sense of what we observe in the natural world," Tanner said. "Theories are structures of ideas that explain and interpret facts."
- Read more about writing a hypothesis, from the American Medical Writers Association.
- Find out why a hypothesis isn't always necessary in science, from The American Biology Teacher.
- Learn about null and alternative hypotheses, from Prof. Essa on YouTube .
Encyclopedia Britannica. Scientific Hypothesis. Jan. 13, 2022. https://www.britannica.com/science/scientific-hypothesis
Karl Popper, "The Logic of Scientific Discovery," Routledge, 1959.
California State University, Bakersfield, "Formatting a testable hypothesis." https://www.csub.edu/~ddodenhoff/Bio100/Bio100sp04/formattingahypothesis.htm
Karl Popper, "Conjectures and Refutations," Routledge, 1963.
Price, P., Jhangiani, R., & Chiang, I., "Research Methods of Psychology — 2nd Canadian Edition," BCcampus, 2015.
University of Miami, "The Scientific Method" http://www.bio.miami.edu/dana/161/evolution/161app1_scimethod.pdf
William M.K. Trochim, "Research Methods Knowledge Base," https://conjointly.com/kb/hypotheses-explained/
University of California, Berkeley, "Multiple Hypothesis Testing and False Discovery Rate" https://www.stat.berkeley.edu/~hhuang/STAT141/Lecture-FDR.pdf
University of California, Berkeley, "Science at multiple levels" https://undsci.berkeley.edu/article/0_0_0/howscienceworks_19
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
What's the difference between a rock and a mineral?
Earth from space: Mysterious, slow-spinning cloud 'cyclone' hugs the Iberian coast
4,000-year-old 'Seahenge' in UK was built to 'extend summer,' archaeologist suggests
Most Popular
- 2 Rare fungal STI spotted in US for the 1st time
- 3 James Webb telescope finds carbon at the dawn of the universe, challenging our understanding of when life could have emerged
- 4 Neanderthals and humans interbred 47,000 years ago for nearly 7,000 years, research suggests
- 5 Noise-canceling headphones can use AI to 'lock on' to somebody when they speak and drown out all other noises
- 2 Bornean clouded leopard family filmed in wild for 1st time ever
- 3 What is the 3-body problem, and is it really unsolvable?
- 4 7 potential 'alien megastructures' spotted in our galaxy are not what they seem
- Current Students
- Online Only Students
- Faculty & Staff
- Parents & Family
- Alumni & Friends
- Community & Business
- Student Life
- Coles College of Business
- Michael J Coles
- Hall of Fame
- Building Bold Connections Podcast
- Rankings & Distinctions
- AACSB Accreditation Information
- Faculty and Staff
- Center for Student Success
- Academic Advising
- Education Abroad
- Internships & Co-ops
- School of Accountancy
- Michael A. Leven School of Management, Entrepreneurship and Hospitality
- Department of Economics, Finance & Quantitative Analysis
- Department of Information Systems and Security
- Department of Marketing & Professional Sales
- Undergraduate
- Executive Education
- All-Access Business Series
- Lessons in Leadership
- Tetley Distinguished Leader Lecture Series
- Faculty Blogs
How does Services Development Affect Manufacturing Export Competitiveness?
KENNESAW, Ga. | Jun 11, 2024
Xuepeng Liu The Research Brief
Most manufacturing activities use service inputs such as financial and business services. Dr. Xuepeng Liu’s paper [1] examines the implications of services development for the export performance of manufacturing sectors. They develop a methodology to quantify the indirect role of services in international trade in goods and construct new measures of revealed comparative advantage based on value-added exports. They show that the development of financial and business services enhances the revealed comparative advantage of manufacturing sectors that use these services intensively but not of other manufacturing sectors. They also find that a country can partially overcome the handicap of an underdeveloped domestic services sector by relying more on imported services inputs. Thus, lower services trade barriers in developing countries can help to promote their manufacturing exports.
The Main Hypothesis
On the face of it, services play a relatively small role in international trade. Conventional trade statistics show that services trade currently accounts for only one-fifth of cross-border trade. However, a significant part of goods trade includes trade in embodied services. In the United States, for example, more than a quarter of intermediate inputs purchased by manufacturers were from the services sector. For certain manufacturing sectors, such as computers and electronic products, this percentage — a measure of “services intensity” — is as high as 47.6 percent. The development of the domestic services sector, as well as access to imported services inputs, can, therefore, be expected to influence comparative advantage in manufacturing trade. Dr. Liu seeks to understand this indirect role of services development drawing upon new measures based on newly available data.
The impact of services development is not straightforward. On the one hand, as services are used as inputs in the production of manufactured goods, services development can help to increase manufacturing production. On the other hand, since services and manufacturing compete for resources, the development of the former can be at the expense of the latter. For example, it is evident that the development of the services sector has drawn resources away from manufacturing not just in developed countries like the United States and the United Kingdom, but also in developing countries like India and the Philippines, provoking “deindustrialization” concerns.
The first hypothesis is that, while the overall effect of services development on the performance of manufacturing sectors is ambiguous, the effect is more likely to be positive for manufacturing sectors that use the services inputs more intensively.
This paper focuses on two services sectors that are crucial for modern economic development: financial services and business services. Both have emerged as skill-intensive, dynamic, internationally traded services. These two services sectors are often regarded as the pillars of modern economies. Services development is mainly measured by the share of a country’s services value-added in GDP. Dr. Liu and his coauthors develop a methodology to quantify the indirect role of services in international trade in goods. They use a suitably modified version of revealed comparative advantage (RCA) to measure the competitiveness of manufacturing sectors. They improve on the traditional Balassa (1965) [2] RCA and construct new measures of RCA based on value-added exports rather than gross exports. Their econometric analysis provides a strong support for the hypothesis.
Policy Implications
Industrial countries have been strong in exporting services, both directly and indirectly. For example, the U.S. is not only the largest direct exporter of business services in the world, but also the largest indirect exporter of business services (actually twice as large), suggesting an important role of business services in U.S.’ manufacturing activities. However, developing and emerging economies have significantly lagged behind, with India being the only exception as a significant direct exporter of business services. Services development in these latter countries would not only strengthen their service sectors but also promote manufacturing sectors.
Countries such as China that may be concerned with the durability of their manufacturing export success may consider building stronger service sectors as a way to upgrade their manufacturing sectors to an even higher level of sophistication. China’s business services exports in value-added terms, relative to its exports in gross terms, are less impressive. Miroudot and Cadestin (2017) [3] show that China is the only country in their sample which has a majority of the manufacturing firms (77 percent in 2013) selling only goods, with little bundling of goods and services, as seen with Apple iPhones/iPads and Apple Stores. To strengthen the manufacturing sector, countries may need to have a favorable business environment that facilitates services upgrading, including but not limited to R&D, marketing, advertising, inventory management, quality control, production scheduling, and after-sale customer services.
With significant improvement in transportation and communication technologies and increasing services outsourcing activities, some developing countries such as India have developed competitive services sectors. For developed countries that have the same strength in service sectors as India, this paper suggests that the manufacturing sectors that use these services intensively tend to have a strong revealed comparative advantage. However, different from most of the other countries, Indian gross exports of business services are actually larger than its total value-added exports, suggesting relatively little embodied business services in other sectors. There is a plenty of room left for India, Philippines and other similar countries to take advantage of their competitive services sectors during their industrialization process.
The Second Hypothesis
This paper also provides evidence for a bypass effect, that is, countries may bypass their inefficient domestic services sectors by making use of imported services inputs. This suggests that nations with under-developed services may take advantage of globalization in services. Countries that hesitate to liberalize their services sectors in the hope of protecting their inefficient domestic services sectors may hurt the competitiveness of their manufacturing sectors.
[1] Liu, Xuepeng, Aaditya Mattoo, Zhi Wang, and Shang-Jin Wei, 2020. "Services Development and Comparative Advantage in Manufacturing.” Journal of Development Economics 144(C) . https://doi.org/10.1016/j.jdeveco.2019.102438
[2] Balassa, Bela, 1965. “Trade Liberalization and ‘Revealed’ Comparative Advantage.” Manchester School of Economic and Social Studies 33: 99-123.
[3] Miroudot, Sébastien, and Charles Cadestin, 2017. “Services In Global Value Chains: From Inputs to Value-Creating Activities.” OECD Trade Policy Papers , No. 197.
Related Posts

The Problem of Teaching Performance Evaluation and a Proposed Solution

Disruption at Regional Universities: Challenges and Opportunities.

Bridging the Gap between Theory and Practice Through Design Science Research

Superintelligence, Conscious Empathic AI, and the Future of Business Research
Contact Info
Kennesaw Campus 1000 Chastain Road Kennesaw, GA 30144
Marietta Campus 1100 South Marietta Pkwy Marietta, GA 30060
Campus Maps
Phone 470-KSU-INFO (470-578-4636)
kennesaw.edu/info
Media Resources
Resources For
Related Links
- Financial Aid
- Degrees, Majors & Programs
- Job Opportunities
- Campus Security
- Global Education
- Sustainability
- Accessibility
470-KSU-INFO (470-578-4636)
© 2024 Kennesaw State University. All Rights Reserved.
- Privacy Statement
- Accreditation
- Emergency Information
- Report a Concern
- Open Records
- Human Trafficking Notice
- Open access
- Published: 13 June 2024

Dyport: dynamic importance-based biomedical hypothesis generation benchmarking technique
- Ilya Tyagin 1 &
- Ilya Safro 2
BMC Bioinformatics volume 25 , Article number: 213 ( 2024 ) Cite this article
Metrics details
Automated hypothesis generation (HG) focuses on uncovering hidden connections within the extensive information that is publicly available. This domain has become increasingly popular, thanks to modern machine learning algorithms. However, the automated evaluation of HG systems is still an open problem, especially on a larger scale.
This paper presents a novel benchmarking framework Dyport for evaluating biomedical hypothesis generation systems. Utilizing curated datasets, our approach tests these systems under realistic conditions, enhancing the relevance of our evaluations. We integrate knowledge from the curated databases into a dynamic graph, accompanied by a method to quantify discovery importance. This not only assesses hypotheses accuracy but also their potential impact in biomedical research which significantly extends traditional link prediction benchmarks. Applicability of our benchmarking process is demonstrated on several link prediction systems applied on biomedical semantic knowledge graphs. Being flexible, our benchmarking system is designed for broad application in hypothesis generation quality verification, aiming to expand the scope of scientific discovery within the biomedical research community.
Conclusions
Dyport is an open-source benchmarking framework designed for biomedical hypothesis generation systems evaluation, which takes into account knowledge dynamics, semantics and impact. All code and datasets are available at: https://github.com/IlyaTyagin/Dyport .
Peer Review reports
Introduction
Automated hypothesis generation (HG, also known as Literature Based Discovery, LBD) has gone a long way since its establishment in 1986, when Swanson introduced the concept of “Undiscovered Public Knowledge” [ 1 ]. It pertains to the idea that within the public domain, there is a significant abundance of information, allowing for the uncovering of implicit connections among various pieces of information. There are many systems developed throughout the years, which incorporate different reasoning methods: from concept co-occurrence in scientific literature [ 2 , 3 ] to the advanced deep learning-based algorithms and generative models (such as BioGPT [ 4 ] and CBAG [ 5 ]). Examples include but are not limited to probabilistic topic modeling over relevant papers [ 6 ], semantic inference [ 7 ], association rule discovery [ 8 ], latent semantic indexing [ 9 ], semantic knowledge network completion [ 10 ] or human-aware artificial intelligence [ 11 ] to mention just a few. The common thread running through these lines of research is that they are all meant to fill in the gaps between pieces of existing knowledge.
The evaluation of HG is still one of the major problems of these systems, especially when it comes to fully automated large-scale general purpose systems (such as IBM Watson Drug Discovery [ 12 ], AGATHA [ 10 ] or BioGPT [ 4 ]). For these, a massive assessment (that is normal in the machine learning and general AI domains) performed manually by the domain experts is usually not feasible and other methods are required.
One traditional evaluation approach is to make a system “rediscover” some of the landmark findings, similar to what was done in numerous works replicating well-known connections, such as: Fish Oil \(\leftrightarrow\) Raynaud’s Syndrome [ 13 ], Migraine \(\leftrightarrow\) Magnesium [ 13 ] or Alzheimer \(\leftrightarrow\) Estrogen [ 14 ]. This technique is frequently used even in a majority of the recently published papers, despite of its obvious drawbacks, such as very limited number of validation samples and their general obsolesce (some of these connections are over 30 years old). Furthermore, in some of these works, the training set is not carefully chosen to include only the information published prior the discovery of interest which turns the HG goal into the information retrieval task.
Another commonly used technique is based on the time-slicing [ 10 , 15 ], when a system is trained on a subset of data prior to a specified cut-off date and then evaluated on the data from the future. This method addresses the weaknesses of previous approach and can be automated, but it does not immediately answer the question of how significant or impactful the connections are. The lack of this information may lead to deceiving results: many connections, even recently published, are trivial (especially if they are found by the text mining methods) and do not advance the scientific field in a meaningful way.
A related area that faces similar evaluation challenges is Information Extraction (IE), a field crucial to enabling effective HG by identifying and categorizing relevant information in publicly available data sources. Within the realm of biomedical and life sciences IE, there are more targeted, small-scale evaluation protocols such as the BioCreative competitions [ 16 ], where the domain experts provide curated training and test datasets, which allows participants to refine and assess their systems within a controlled environment. While such targeted evaluations as conducted in BioCreative are both crucial and insightful, they inherently lack the scope and scale needed for the evaluation of expansive HG systems.
The aforementioned issues emphasize the critical need for research into effective, scalable evaluation methods in automated hypothesis generation. Our primary interest is in establishing an effective and sustainable benchmark for large-scale, general-purpose automated hypothesis generation systems within the biomedical domain. We seek to identify substantial, non-trivial insights, prioritizing them over mere data volume and ensuring scalability with respect to ever-expanding biocurated knowledge databases. We emphasize the significance of implementing sustainable evaluation strategies, relying on constantly updated datasets reflecting the latest research. Lastly, our efforts are targeted towards distinguishing between hypotheses with significant impact and those with lesser relevance, thus moving beyond trivial generation of hypotheses to ensuring their meaningful contribution to scientific discovery.
Our contribution
We propose a high quality benchmark dataset Dyport for hypothesis prediction systems evaluation. It incorporates information extracted from a number of biocurated databases. We normalize all concepts to the unified format for seamless integration and each connection is supplied with rich metadata, including timestamp information to enable time-slicing.
We introduce an evaluation method for the impact of connections in time-slicing paradigm. It will allow to benchmark HG systems more thoroughly and extensively by assigning an importance weight to every connection over the time. This weight represents the overall impact a connection makes on future discoveries.
We demonstrate the computational results of several prediction algorithms using the proposed benchmark and discuss their performance and quality.
We propose to use our benchmark to evaluate the quality of HG systems. The benchmark is designed to be updated on a yearly basis. Its structure facilitates relatively effortless expansion and reconfiguration by users and developers.
Background and related work
Unfortunately, the evaluation in the hypothesis generation field is often coupled with the systems to evaluate and currently not universally standardized. If one would like to compare the performance of two or more systems, they need to understand their training protocol to instantiate models from scratch and then test them on the same data they used in their experiment.
This problem is well known and there are attempts to provide a universal way to evaluate such systems. For example, OpenBioLink [ 17 ] is designed as a software package for evaluation of link prediction models. It supports time-slicing and contains millions of edges with different quality settings. The authors describe it as “highly challenging” dataset that does not include “trivially predictable” connections, but they do not provide a quantification of difficulty nor range the edges accordingly.
Another attempt to set up a large-scale validation of HG systems was performed in our earlier work [ 18 ]. The proposed methodology is based on the semantic triples extracted from SemMedDB [ 19 ] database and setting up a cut date for training and testing. Triples are converted to pairs by removing the “verb” part from each (subject-verb-object) triple. For the test data, a list of “highly cited” pairs is identified, which is based on the citation counts from SemMedDB, MEDLINE and Semantic Scholar. Only connections occurring in papers published after the cut date and cited over 100 times are considered. It is worth mentioning that this approach is prone to noise (due to SemMedDB text mining methods) and also skewed towards the discoveries published closer to the cut-date, since the citations accumulate over time.
One more aspect of the proposed approach relates to the quantification and detection of scientific novelty. Research efforts range from protein design domain studies [ 20 ] to analyzing scientific publications through their titles [ 21 ] or using manual curation in combination with machine learning [ 22 ]. However, none of these techniques were integrated into a general purpose biomedical evaluation framework, where the novelty would be taken into account.
Currently, Knowledge Graph Embeddings (KGE) are becoming increasingly popular and the hypothesis generation problem can be formulated in terms of link prediction in knowledge graphs. Knowledge Graphs often evaluate the likelihood of a particular connection with the scoring function of choice. For example, TransE [ 23 ] evaluates each sample with the following equation:
where h is the embedding vector of a head entity, r is the embedding vector of relation, t is the embedding vector of a tail entity and \(||\cdot ||\) denotes the L1 or L2 norm.
These days KGE-based models are of interest to the broad scientific community, including researchers in the drug discovery field. Recently they carefully investigated the factors affecting the performance of KGE models [ 24 ] and reviewed biomedical databases related to drug discovery [ 25 ]. These publications, however, do not focus on any temporal information nor attempt to describe the extracted concept associations quantitatively. We also aim to fill in this currently existing gap in our current work.
\(c_i\) —concept in some arbitrary vocabulary;
\(m(\cdot )\) —function that maps a concept \(c_i\) to the subset of corresponding UMLS CUI. The result is denoted by \(m_i =m(c_i)\) . The \(m_i\) is not necessarily a singleton. We will somewhat abuse the notation by denoting \(m_i\) a single or any of the UMLS terms obtained by mapping \(c_i\) to UMLS.
\(m(\cdot ,\cdot )\) —function that maps pairs of \(c_i\) and \(c_j\) into the corresponding set of all possible UMLS pairs \(m_i\) and \(m_j\) . Recall that the mapping of \(c_i\) to UMLS may not be unique. In this case \(|m(c_i,c_j)| = |m(c_i)|\cdot |m(c_j)|\) .
\((m_i, m_j)\) —a pair of UMLS CUIs, which is extracted as a co-occurrence from MEDLINE records. It also represents an edge in network G and is cross-referenced with biocurated databases;
D —set of pairs \((m_i, m_j)\) extracted from biocurated databases;
P —set of pairs \((m_i, m_j)\) extracted from MEDLINE abstracts;
E —set of cross-referenced pairs \((m_i, m_j)\) , such that \(E = D \cap P\) ;
G —dynamic network, containing temporal snapshots \(G_t\) , where t —timestamp (year);
\(\hat{G}_t\) —snapshot of network G for a timestamp t only containing nodes from \(G_{t-1}\) .
The main unit of analysis in HG is a connection between two biomedical concepts, which we also refer to as “pair”, “pairwise interaction” or “edge” (in network science context when we will be discussing semantic networks). These connections can be obtained from two main sources: biomedical databases and scientific texts. Extracting pairs from biomedical databases is done with respect to the nature and content of the database: some of them already contain pairwise interactions, whereas others focus on more complex structures such as pathways which may contain multiple pairwise interactions or motifs (e.g., KEGG [ 26 ]). Extracting pairs from textual data is done via information retrieval methods, such as relation extraction or co-occurrence mining. In this work, we use the abstract-based co-occurrence approach, which is explained later in the paper.
Method in summary
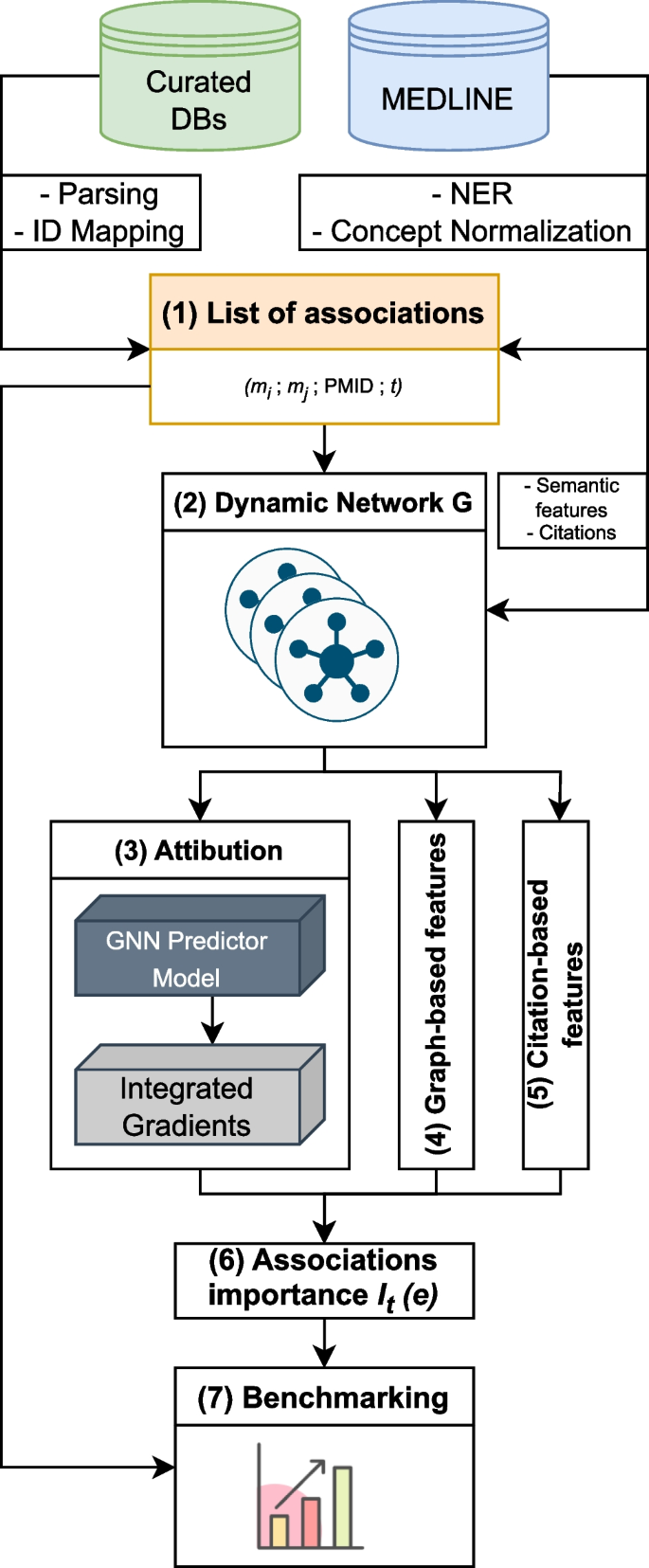
Summary of the HG benchmarking approach. We start with collecting data from Curated DBs and Medline, then process it: records from Curated DBs go through parsing, cleaning and ID mapping, MEDLINE records are fed into SemRep system, which performs NER and concept normalization. After that we obtain a list of UMLS CUI associations with attached PMIDs and timestamps (TS). This data is then used to construct a dynamic network G , which is used to calculate the importance measure I for edges in the network. At the end, edges \(e \in G\) with their corresponding importance scores \(I_t(e)\) are added to the benchmark dataset
The HG benchmarking pipeline is presented in Fig. 1 . The end goal of the pipeline is to provide a way to evaluate any end-to-end hypothesis generation system trained to predict potential pairwise associations between biomedical instances or concepts.
We start with collecting pairwise entity associations from a list of biocurated databases, which we then normalize and represent as pairs of UMLS [ 27 ] terms \((m_i, m_j)\) . The set of these associations is then cross-referenced with scientific abstracts extracted from MEDLINE database, such that for each pair \((m_i, m_j)\) we keep all PubMed identifiers (PMID) that correspond to the paper abstracts in which \(m_i\) and \(m_j\) co-occured. As a result, there is a list of tuples (step 1, Fig. 1 ) \((m_i, m_j, \text {PMID}, t)\) , where t is a timestamp for a given PMID extracted from its metadata. We then split this list into a sequence \(\{E_t\}\) according to the timestamp t . In this work t is taken with a yearly resolution.
Each individual \(E_t\) can be treated as an edgelist, which yields an edge-induced network \(G_t\) constructed from edges \((m_i, m_j) \in E_t\) . It gives us a sequence of networks \(G = \{G_t\}\) (step 2, Fig. 1 ), which is then used to compute the importance of individual associations in \(E_t\) with different methods.
The main goal of importance is to describe each edge from \(E_t\) using additional information. The majority of it comes from the future network snapshot \(G_{t+1}\) , which allows us to track the impact that a particular edge had on the network in the future. The predictive impact is calculated with an attribution technique called Integrated Gradients (IG) (step 3, Fig. 1 ). Structural impact is calculated with graph-based measures (such as centrality) (step 4, Fig. 1 ) and citation impact is calculated with respect to how frequently edges are referenced in the literature after their initial discovery (step 5, Fig. 1 ).
All the obtained scores are then merged together to obtain a ranking \(I_t(e)\) (step 6, Fig. 1 ), where \(e \in E_t\) for all edges from a snapshot \(G_t\) . Finally, this ranking is used to perform stratified evaluation of how well hypothesis generation systems perform in discovering connections with different importance values (step 7, Fig. 1 ).
Databases processing and normalization
We begin by gathering the links and relationships from publicly available databases, curated by domain experts. We ensure that all pairwise concept associations we utilize are from curated sources. For databases like STRING, which compile associations from various channels with differing levels of confidence, we exclusively select associations derived from curated sources.
Ensuring correct correspondence of the same concepts from diverse databases is highly crucial. Therefore, we also conduct mapping of all concepts to UMLS CUI (Concept Unique Identifier). Concepts, which identifiers cannot be mapped to UMLS CUI, are dropped. In our process, we sometimes encounter situations where a concept \(c_{i}\) , may have multiple mappings to UMLS CUIs, i.e., \(|m_i|=k>1\) for \(m_i = m(c_i)\) . To capture these diverse mappings, we use the Cartesian product rule. In this approach, we take the mapping sets for both concepts \(c_{i}\) and \(c_{j}\) , denoted as \(m(c_{i})\) and \(m(c_{j})\) , and generate a new set of pairs encapsulating all possible combinations of these mappings. Essentially, for each original pair \((c_{i}, c_{j})\) , we produce a set of pairs \(m(c_{i}, c_{j})\) such that the cardinality of this new set equals the product of the cardinalities of the individual mappings. Let us say that \(c_i\) has k different UMLS mappings and \(c_j\) has s , then \(|m(c_{1},c_{2})| = |m(c_{1})| \cdot |m(c_{2})| = k\cdot s\) .
In other words, we ensure that every possible mapping of the original pair is accounted for, enabling our system to consider all potential pairwise interactions across all UMLS mappings. To this end, we have collected all pairs of UMLS CUI that are present in different datasets, forming a set D .
Processing MEDLINE records
To match pairwise interactions extracted from biocurated databases to literature, we use records from MEDLINE database with their PubMed identifiers. These records, primarily composed of the titles and abstracts of scientific papers, are each assigned a unique PubMed reference number (PMID). They are also supplemented with rich metadata, which includes information about authors, full-text links (when applicable), and date of publication timestamps indicating when the record became publicly available. We process records with an NLM-developed natural language processing tool SemRep [ 28 ] to perform named entity recognition, concept mapping and normalization. To this end, we obtain a list of UMLS CUI for each MEDLINE record.
Connecting database records with literature
The next step is to form connections between biocurated records and their corresponding mentions in the literature. With UMLS CUIs identified in the previous step, we track the instances where these CUIs are mentioned together within the same scientific abstract. Our method considers the simultaneous appearance of a pair of concepts, denoted as \(m_i\) and \(m_j\) , within a single abstract to represent a co-occurrence. This co-occurrence may indicate a potential relationship between the two concepts within the context of the abstract. All the co-occurring pairs \((m_i, m_j)\) , extracted from MEDLINE abstracts, form the set P .
No specific “significance” score is assigned to these co-occurrences at this point beyond their presence in the same abstract. Subsequently, these pairs are cross-referenced with pairs in biocurated databases. More specifically, for each co-occurrence \((m_i, m_j) \in P\) we check its presence in set D . Pairs not present in both sets D and P are discarded. This forms the set E :
This step validates each co-occurring pair, effectively reducing noise and confirming that each pair holds biological significance. Conversely, E can be described as a set of biologically relevant associations, with each element enriched by contextual information extracted from scientific literature. The procedure is described in [ 29 ] as distant supervised annotation .
Constructing time-sliced graphs
After we find the set of co-occurrences in abstracts extracted from MEDLINE and cross-referenced with pairs in biocurated databases (set E ), we split it based on the timestamps extracted from the abstracts metadata. The timestamps t are assigned to each PMID and are used to determine when they became publicly available. We use these timestamps to track how often was a pair of UMLS CUIs \((m_i, m_j)\) appearing in the biomedical literature over time. As a result, we have a list of biologically relevant cross-referenced UMLS CUI co-occurrences, each connected to all PMIDs containing them.
This list is then split into edge lists \(E_t\) , such that each edge list contains pairs \((m_i, m_j)\) added in or before year t . These edge lists are then transformed to dynamic network G with T snapshots:
where \(N_t\) and \(E_t\) represent the set of unique UMLS CUIs (nodes) and their cross-referenced abstract co-occurrences (edges), respectively, and t is the annual timestamp (time resolution can be changed as needed), such that \(G_{t}\) is constructed from all MEDLINE records published before t (e.g., \(t=2011\) ). All networks \(G_{t}\) are simple and undirected.
For each timestamp t , \(G_{t}\) represents a cumulative network, including all the information from \(G_{t-1}\) and new information added in year t .
Tracking the edge importance of time-sliced graphs
We enrich the proposed benchmarking strategy with the information about associations importance at each time step t . In the context of scientific discovery, the importance may be considered from several different perspectives, e.g., as an the influence of an individual finding on future discoveries. In this section we take three different perspectives into account and then combine them together to obtain a final importance score, which we later use to evaluate different hypothesis generation systems with respect to their ability to predict the important associations.
Integrated gradients pipeline
In this step we obtain the information about how edges from graph \(G_t\) influence the appearance of new edges in \(G_{t+1}\) . For that we train a machine learning model, which is able to perform link predictions and then we use an attribution method called Integrated Gradients (IG).
In general, IG is used to understand input features importance with respect to the output a given predictor model produces. In case of link prediction problem, a model outputs likelihood of two nodes \(m_i\) and \(m_j\) being connected for a given network \(G_t\) . The input features for a link prediction model will include the adjacency matrix of \(G_t\) , \(A_t\) , and the predictions themselves can be drawn from a list of edges appearing in the next timestamp \(t + 1\) . If IG is applied to this particular problem, it will provide attribution values for each element of \(A_t\) , which can be reformulated as the importance of edges existing at the timestamp t with respect to their contribution to predicting the edges added at the next timestamp \(t+1\) . This could be interpreted as the influence of current dynamic network structural elements on the information that will be added in future.
Link prediction problem In our setting, the link prediction problem is formulated as following:
We note that predictions of edges \(\hat{E}_{t+1}\) are performed only for nodes \(N_t\) from the graph \(G_t\) at year t .
Adding Node and Edge Features : To enrich the dynamic network G with non-redundant information extracted from text, we add node features and edge weights. Node features are required for Graph Neural Network-based predictor training, which we use in the proposed pipeline.
Node features : Node features are added to each \(G_t\) by applying word2vec algorithm [ 30 ] to the corresponding snapshot of MEDLINE dataset obtained for a timestamp t . In order to perform cleaning and normalization, we replace all tokens in the input texts by their corresponding UMLS CUIs obtained at the NER stage. It significantly reduces the vocabulary size, automatically removing stop-words and enabling vocabulary-guided phrase mining [ 31 ]. It is important to note that each node m has a different vector representation for each time stamp t , which we can refer to as n 2 v ( m , t ).
Edge features (weights) : For simplicity, edge weights are constructed by counting the number of MEDLINE records mentioning a pair of concepts \(e \in E_{t}\) . In other words, for each pair \(e = (m_i, m_j)\) we assign a weight representing the total number of mentions for a pair e in year t .
GNN training
We use a graph neural network-based encoder-decoder architecture. Its encoder consists of two graph convolutional layers [ 32 ] and produces an embedding for each graph node. Decoder takes the obtained node embeddings and outputs the sum of element-wise multiplication of encoded node representations as a characteristic of each pair of nodes.
Attribution
To obtain a connection between newly introduced edges \(\hat{E}_{t+1}\) and existing edges \(E_t\) , we use an attribution method Integrated Gradients (IG) [ 33 ]. It is based on two key assumptions:
Sensitivity: any change in input that affects the output gets a non-zero attribution;
Implementation Invariance: attribution is consistent with the model’s output, regardless of the model’s architecture.
The IG can be applied to a wide variety of ML models as it calculates the attribution scores with respect to input features and not the model weights/activations, which is important, because we focus on relationships between the data points and not the model internal structure.
The integrated gradient (IG) score along \(i^{th}\) dimension for an input x and baseline \(x'\) is defined as:
where \(\frac{\partial F(x)}{\partial x_i}\) is the gradient of F ( x ) along \(i^{th}\) dimension. In our case, input x is the adjacency matrix of \(G_t\) filled with 1 s as default values (we provide all edges \(E_t \in G_t\) ) and baseline \(x'\) is the matrix of zeroes. As a result, we obtain an adjacency matrix \(A(G_t)\) filled with attribution values for each edge \(E_t\) .
Graph-based measures
Betweenness Centrality In order to estimate the structural importance of selected edges, we calculate their betweenness centrality [ 34 ]. This importance measure shows the amount of information passing through the edges, therefore indicating their influence over the information flow in the network. It is defined as
where \(\sigma _{st}\) —the number of shortest paths between nodes s and t ; \(\sigma _{st}(e)\) —the number of shortest paths between nodes s and t passing through edge e .
To calculate the betweenness centrality with respect to the future connections, we restrict the set of vertices V to only those, that are involved in future connections we would like to use for explanation.
Eigenvector Centrality Another graph-based structural importance metric we use is the eigenvector centrality. The intuition behind it is that a node of the network is considered important if it is close to other important nodes. It can be found as a solution of the eigenvalue problem equation:
where A is the network weighted adjacency matrix. Finding the eigenvector corresponding to the largest eigenvalue gives us a list of centrality values \(C_E(v)\) for each vertex \(v \in V\) .
However, we are interested in edge-based metric, which we obtain by taking an absolute difference between the adjacent vertex centralities:
where \(e=(u,v)\) . The last step is to connect this importance measure to time snapshot, which we do by taking a time-base difference between edge-based eigenvector centralities
This metric gives us the eigenvector centrality change with respect to future state of the dynamic graph ( \(t+1\) ).
Second Order Jaccard Similarity One more indicator of how important a particular newly discovered network connection is related to its adjacent nodes neighborhood similarity. The intuition is that more similar their neighborhood is, more trivial the connection is, therefore, it is less important.
We consider a second-order Jaccard similarity index for a given pair of nodes \(m_i\) and \(m_j\) :
Second-order neighborhood of a node u is defined by:
where w iterates over all neighbors of u and N ( w ) returns the neighbors of w .
The second order gives a much better “resolution” or granularity for different connections compared to first-order neighborhood. We also note that it is calculated for a graph \(G_{t-1}\) for all edges \(\hat{E}_{t}\) (before these edges were discovered).
Literature-based measures
Cumulative citation counts Another measure of a connection importance is related to bibliometrics. At each moment in time for each targeted edge we can obtain a list of papers mentioning this edge.
We also have access to a directed citation network, where nodes represent documents and edges represent citations: edges connect one paper to all the papers that it cites. Therefore, the number of citations of a specific paper would equal to in-degree of a corresponding node in a citation network.
To connect paper citations to concepts connections, we compute the sum of citation counts of all papers mentioning a specific connection. Usually, the citation counts follow heavy-tailed distributions (e.g., power law) and counting them at the logarithmic scale is a better practice. However, in our case the citation counts are taken “as-is” to emphasize the difference between the number of citations and the number of mentions. This measure shows the overall citation-based impact of a specific edge over time. The citation information comes from the citation graph, which is consistent with the proposed dynamic network in terms of time slicing methodology.
Combined importance measure for ranking connections
To connect all the components of the importance measure I for edge e , we use the mean percentile rank (PCTRank) of each individual component:
where \(C_i\) is the importance component (one of the described earlier, C —set of all importance components). The importance measure is calculated for each individual edge in graph for each moment in time t with respect to its future (or previous) state \(t+1\) (or \(t-1\) ). Using the mean percentile rank guarantees that the component will stay within a unit interval. The measure I is used to implement an importance-based stratification strategy for benchmarking, as it is discussed in Results section.
In this section we describe the experimental setup and propose a methodology based on different stratification methods. This methodology is unique for the proposed benchmark, because each record is supplied with additional information giving a user more flexible evaluation protocol.
Data collection and processing
Dynamic graph construction.
The numbers of concepts and their associations successfully mapped to UMLS CUI \((m_i, m_j)\) from each dataset are summarized in Table 1 . The number of associations with respect to time is shown in Fig. 2 . It can be seen that the number of concept associations steadily and consistently grows for every subsequent year.
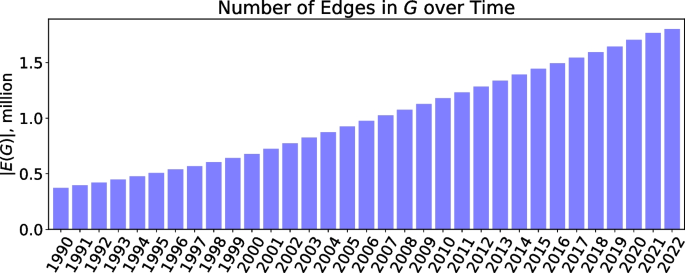
Number of edges in the network G over time. The numbers are reported in millions. Each edge represents a pair of cross-referenced UMLS CUI concepts \((m_i, m_j)\)
Data collection and aggregation is performed in the following pipeline:
All databases are downloaded in their corresponding formats such as comma-separated or Excel spreadsheets, SQL databases or Docker images.
All pairwise interactions in each database are identified.
From all these interactions we create a set of unique concepts, which we then map to UMLS CUIs. Concepts that do not have UMLS representations are dropped.
All original pairwise interactions are mapped with respect to the UMLS codes, as discussed in Databases Processing and Normalization section.
A set of all pairwise interactions is created by merging the mapped interactions from all databases.
This set is then used to find pairwise occurrences in MEDLINE.
Pairwise occurrences found in step 6 are used to construct the main dynamic network G . As it was mentioned earlier, G is undirected and non-attributed (we do not provide types of edges as they are much harder to collect reliably on large scale), which allows us to cover a broader range of pairwise interactions and LBD systems to test. Other pairwise interactions, which are successfully mapped to UMLS CUI, but are not found in the literature, can still be used. They do not have easily identifiable connections to scientific literature and do not contain temporal information, which make them a more difficult target to predict (will be discussed later).
Compound importance calculation
Once the dynamic graph G is constructed, we calculate the importance measure. For that we need to decide on three different timestamps:
Training timestamp: when the predictor models of interest are trained;
Testing timestamp: what moment in time to use to accumulate recently (with respect to step 1) discovered concept associations for models testing;
Importance timestamp: what moment in time to use to calculate the importance measure for concept associations from step 2.
To demonstrate our benchmark, we experiment with different predictive models. In our experimental setup, all models are trained on the data published prior to 2016, tested on associations discovered in 2016 and the importance measure I is calculated based on the most recent fully available timestamp (2022, at the time of writing) with respect to the PubMed annual baseline release. We note that, depending on the evaluation goals, other temporal splits can be used as well. For example, one can decide to evaluate the predictive performance of selected models on more recently discovered connections. For that, they may use the following temporal split: training timestamp—2020, testing timestamp—2021, importance timestamp—2022.
The importance measure I has multiple components, which are described in Methods section. To investigate their relationships and how they are connected to each other, we plot a Spearman correlation matrix showed in Table 2 . Spearman correlation is used because only component’s rank matters in the proposed measure as all components are initially scaled differently.
Evaluation protocol
In our experiments, we demonstrate a scenario for benchmarking hypothesis generation systems. All of the systems are treated as predictors capable of ranking true positive samples (which come from the dynamic network G ) higher than the synthetically generated negatives. The hypothesis generation problem is formulated as binary classification with significant class imbalance.
Evaluation metric
The evaluation metric of choice for our benchmarking is Receiver Operating Characteristic (ROC) curve and its associated Area Under the Curve (AUC), which is calulated as:
where \({\textbf {1}}\) is the indicator function that equals 1 if the score of a negative example \(t_0\) is less than the score of a positive example \(t_1\) ; \(D^0\) , \(D^1\) are the sets of negative and positive examples, respectively. The ROC AUC score quantifies the model’s ability to rank a random positive higher than a random negative.
We note than the scores do not have to be within a specific range, the only requirement is that they can be compared with each other. In fact, using this metric allows us to compare purely classification-based models (such as Node2Vec logistic regression pipeline) and ranking models (like TransE or DistMult), even though the scores of these models may have arbitrary values.
Negative sampling
Our original evaluation protocol can be found in [ 10 ], which is called subdomain recommendation . It is inspired by how biomedical experts perform large-scale experiments to identify the biological instances of interest from a large pool of candidates [ 35 ]. To summarize:
We collect all positive samples after a pre-defined cut date. The data before this cut date is used for prediction system training.
For each positive sample (subject-object pair) we generate N negative pairs, such that the subject is the same and the object in every newly generated pair has the same UMLS semantic type as the object in positive pair;
We evaluate a selected performance measure (ROC AUC) with respect to pairs of semantic types (for example, gene-gene or drug-disease) to better understand domain specific differences.
For this experiment we set \(N=10\) as a trade-off between the evaluation quality and runtime. It can be set higher if more thorough evaluation is needed.
Baseline models description
To demonstrate how the proposed benchmark can be used to evaluate and compare different hypothesis generation system, we use a set of existing models. To make the comparison more fair, all of them are trained on the same snapshots of MEDLINE dataset.
The AGATHA is a general purpose HG system [ 10 , 36 ] incorporates a multi-step pipeline, which processes the entire MEDLINE database of scientific abstracts, constructs a semantic graph from it and trains a predictor model based on transformer encoder architecture. Besides the algorithmic pipeline, the key difference between AGATHA and other link prediction systems is that AGATHA is an end-to-end hypothesis generation framework, where the link prediction is only one of its components.
Node2Vec-based predictor is trained as suggested in the original publication [ 37 ]. We use a network purely constructed with text-mining-based methods.
Knowledge graph embeddings-based models
Knowledge Graph Embeddings (KGE) models are becoming increasingly popular these days, therefore we include them into our comparison. We use Ampligraph [ 38 ] library to train and query a list of KGE models: TransE, HolE, ComplEx and DistMult.
Evaluation with different stratification
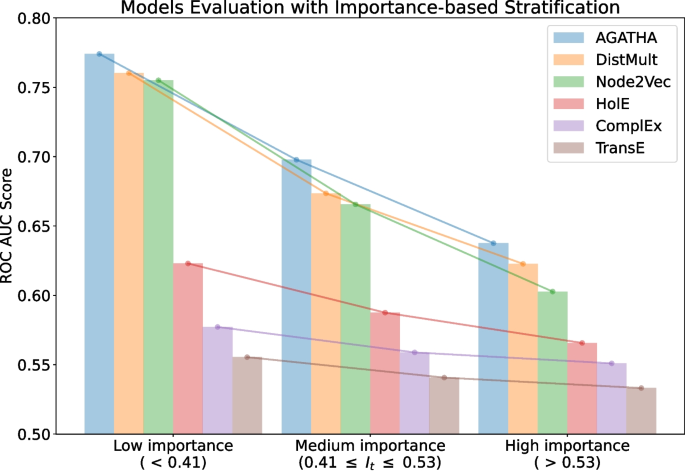
ROC AUC scores for different models trained on the same PubMed snapshot from 2015 and tested on semantic predicates added in 2016 binned with respect to their importance scores
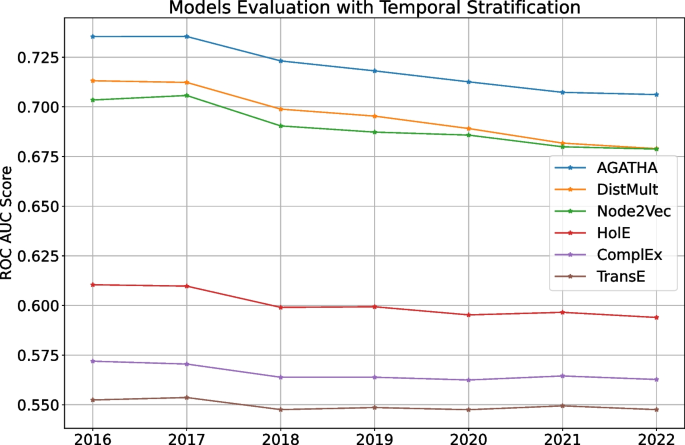
ROC AUC scores for different models trained on the same PubMed snapshot from 2015 and tested on semantic predicates added over time
The proposed benchmarking pipeline enables us to perform different kinds of systems evaluation and comparison with flexibility usually unavailable to other methods. Incorporating both temporal and importance information is helpful to identify trends in models behavior and extend the variety of criteria for domain experts when they decide on a best model suitable for their needs.
Below we present three distinct stratification methods and show how predictor models perform under different evaluation protocols. Even though we use the same performance metric (ROC AUC) across the board, the results differ substantially, suggesting that evaluation strategy plays a significant role in the experimental design.
Semantic stratification
Semantic stratification strategy is the natural way to benchmark hypothesis generation systems, when the goal is to evaluate performance in specific semantic categories. It is especially relevant to the subdomain recommendation problem, which defines our negative sampling procedure. For that we take the testing set of subject-object pairs and group them according to their semantic types and evaluate each group separately (Table 3 ).
Importance-based stratification
The next strategy is based on the proposed importance measure I . This measure ranks all the positive subject-object pairs from the test set and, therefore, can be used to split them into equally-sized bins, according to their importance score. In our experiment, we split the records into three bins, representing low, medium and high importance values. Negative samples are split accordingly. Then each group is evaluated separately. The results of this evaluation are presented in Fig. 3 .
The results indicate that the importance score I could also reflect the difficulty of making a prediction. Specifically, pairs that receive higher importance scores tend to be more challenging for the systems to be identified correctly. In models that generally exhibit high performance (e.g., DistMult), the gap in ROC AUC scores between pairs with low importance scores and those with high importance scores is especially pronounced. The best model in this list is AGATHA as it utilizes the most nuanced hypothesis representation, namely, its transformer architecture is trained to leverage not only node embeddings but also to account for the non-overlapping neighborhoods of concepts.
Temporal stratification
The last strategy shows how different models trained once perform over time . For that we fix the training timestamp on 2015 and evaluate each models on testing timestamps from 2016 to 2022. For clarity, we do not use importance values for this experiment and only focus on how the models perform over time on average . The results are shown in Fig. 4 .
Figure 4 highlights how predictive performance gradually decays over time for every model in the list. This behavior can be expected: the gap between training and testing data increases over time, which makes it more difficult for models to perform well as time goes by. Therefore, it is a good idea to keep the predictor models up-to-date, which we additionally discuss in the next section.
We divide the discussion into separate parts: topics related to evaluation challenges and topics related to different predictor model features. We also describe the challenges and scope for the future work at the end of the section.
Evaluation-based topics
Data collection and processing challenges.
The main challenge of this work comes from the diverse nature of biomedical data. This data may be described in many different ways and natural language may not be the most commonly used. Our results indicate that a very significant part of biocurated connections “flies under the radar” of text-mining systems and pipelines due to several reasons:
Imperfections of text-mining methods;
Multiple standards to describe biomedical concepts;
The diversity of scientific language: many biomedical associations (e.g. gene-gene interactions may be primarily described in terms of co-expression);
Abstracts are not enough for text mining [ 39 ].
The proposed methodology for the most part takes the lowest common denominator approach: we discard concepts not having UMLS representations and associations not appearing in PubMed abstracts. However, our approach still allows us to extract a significant number of concept associations and to use them for quantitative analysis. We should also admit that the aforementioned phenomenon of biomedical data discrepancy leads us to some interesting results, which we discuss below.
Different nature of biomedical DBs and literature-extracted data
The experiment clearly indicates significant differences between different kinds of associations with respect their corresponding data sources in models performance comparison. For this experiment we take one of the evaluated earlier systems (AGATHA 2015) and run the semantically-stratified version of benchmark collected from three different data sources:
Proposed benchmark dataset: concept associations extracted from biocurated databases with cross-referenced literature data;
Concept associations extracted from biocurated databases, but which we could not cross-reference with literature data;
Dataset composed of associations extracted with a text mining framework (SemRep).
Datasets (1) and (3) were constructed from associations found in MEDLINE snapshot from 2020. For dataset (2) it was impossible to identify the time connections were added, therefore the cut date approach was not used. All three datasets were downsampled with respect to the proposed benchmark (1), such that the number of associations is the same across all of them.
The results of this experiment are shown in Table 4 . It is evident that associations extracted from biocurated databases (1) and (2) propose a more significant challenge for a text-mining-based system. Cross-referencing from literature makes sure that similar associations can be at least discovered by these systems at the training time, therefore, the AGATHA performance on dataset (1) is higher compared to dataset (2). These results may indicate that biocurated associations, which cannot be cross-referenced, belong to a different data distribution, and, therefore, purely text mining-based systems fall short due to the limitations of the underlying information extraction algorithms.
Models-related topics
Text mining data characteristics.
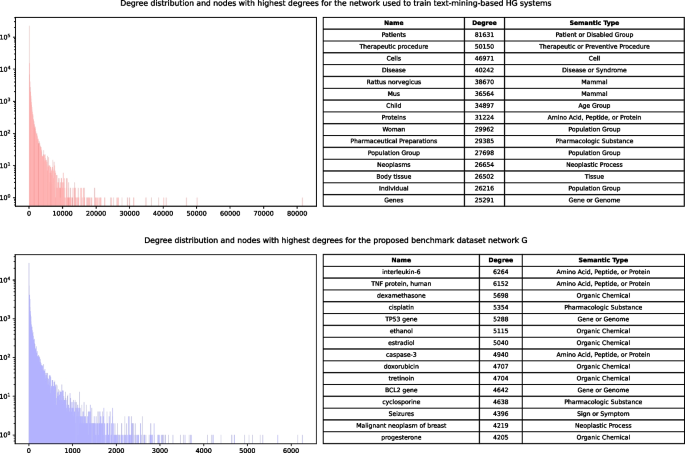
Degree distributions and nodes with highest degrees for two networks: the one used for training of text-mining-based predictor models (red, top) and the network G from the proposed benchmark dataset (blue, bottom)
In order to demonstrate the differences between biologically curated and text mining-based knowledge, we can consider their network representations.
The network-based models we show in this work are trained on text-mining-based networks, which are built on top of semantic predicates extracted from a NLP tool SemRep. This tool takes biomedical text as input and extracts triples (subject-verb-object) from the text and performs a number of additional tasks, such as:
Named Entity Recognition
Concept Normalization
Co-reference Resolution
and some others. This tool operates on UMLS Metathesaurus, one of the largest and most diverse biomedical thesaurus, including many different vocabularies.
The main problem of text-mining tools like SemRep is that they tend to produce noisy (and often not quite meaningful from the biomedical prospective) data. As a result, the underlying data that is used to build and validate literature-based discovery systems may not represent the results that domain experts expect to see.
However, these systems are automated and, therefore, are widely used as a tool to extract information from literature in uninterrupted manner. Then this information is used for training different kinds of predictors (either rule-based, statistical or deep learning).
To demonstrate this phenomenon, we compare two networks, where nodes are biomedical terms and edges are associations between them. The difference between them lies in their original data source, which is either:
PubMed abstracts processed with SemRep tool;
Biocurated databases, which connections are mapped to pairs of UMLS CUI terms and cross-referenced with MEDLINE records.
Connections from the network (2) are used in the main proposed benchmarking framework (network G ). The comparison is shown in Fig. 5 as a degree distribution of both networks. We can see that network (1) has a small number of very high-degree nodes. These nodes may affect negatively to the overall predictive power of any model using networks like (1) as a training set, because they introduce a large number of “shortcuts” to the network, which do not have any significant biological value. We also show the top most high-degree nodes for both networks. For the network (1), all of them appear to be very general and most of them (e.g. “Patients” or “Pharmaceutical Preparations”) can be described as noise. Network (2), in comparison, contain real biomedical entities, which carry domain-specific meaning.
Training data threshold influence
As the Temporal Stratification experiment in the Results section suggests, the gap between training and testing timestamps plays a noticeable role in models predictive performance.
To demonstrate this phenomena from a different perspective, we now fix the testing timestamp and vary the training timestamp. We use two identical AGATHA instances, but trained on different MEDLINE snapshots: 2015 and 2020. The testing timestamp for this experiment is 2021, such that none of the models has access to the test data.
The results shown in Table 5 illustrate that having more recent training data does not significantly increase model’s predictive power for the proposed benchmark. This result may be surprising, but there is a possible explanation: a model learns the patterns from the training data distribution and that data distribution stays consistent for both training cut dates (2015 and 2020). However, that does not mean that the data distribution in the benchmark behaves the same way. In fact, it changes with respect to both data sources: textual and DB-related.
Semantic types role in predictive performance
Another aspect affecting models predictive performance is having access to domain information. Since we formulate the problem as subdomain recommendation, knowing concept-domain relationships may be particularly valuable. We test this idea by injecting semantic types information into the edge type for tested earlier Knowledge Graph Embedding models. As opposed to classic link prediction methods (such as node2vec), Knowledge Graph modeling was designed around typed edges and allows this extension naturally.
Results in Table 6 show that semantic type information provides a very significant improvement for models predictive performance.
Large language models for scientific discovery
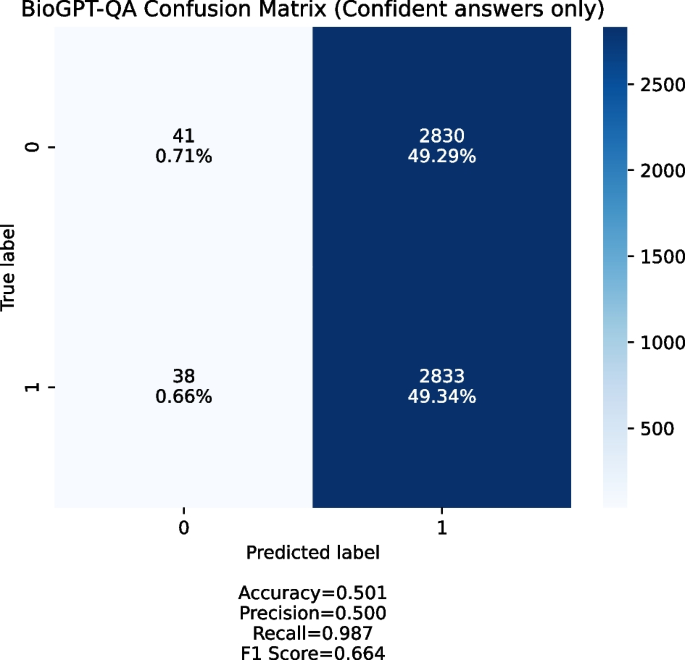
Confusion matrix obtained by the BioGPT-QA model. Only confident answers (Yes/No) were taken into account
Recent advances in language model development raised a logical question about usefulness of these models in scientific discovery, especially in biomedical area [ 40 ]. Problems like drug discovery, drug repurposing, clinical trial optimization and many others may benefit significantly from systems trained on a large amount of scientific biomedical data.
Therefore, we decide to test how one of these systems would perform in our benchmark. We take one of the recently released generative pre-trained transformer models BioGPT [ 4 ] and run a set of test queries.
BioGPT model was chosen due to the following reasons:
It is recently released (2022);
It includes fine-tuned models, which show good performance on downstream tasks;
It is open source and easily accessible.
We use a BioGPT-QA model to perform the benchmarking, because it was fine-tuned on PubMedQA [ 41 ] dataset and outputs the answer as yes/maybe/no, which is easy to parse and represent as a (binary) classifier output.
The question prompt was formulated as the following: “Is there a relationship between <term 1> and <term 2>?”. PubMedQA format also requires a context from a PubMed abstract, which does not exist in our case, because it is a discovery problem. However, we supply an abstract-like context, which is constructed by concatenating term definitions extracted from UMLS Metathesaurus for both source and target terms.
A sample prompt looks like this: “Is there a relationship between F1-ATPase and pyridoxal phosphate? context: F1-ATPase—The catalytic sector of proton-translocating ATPase complexes. It contains five subunits named alpha, beta, gamma, delta and eta. pyridoxal phosphate—This is the active form of VITAMIN B6 serving as a coenzyme for synthesis of amino acids, neurotransmitters (serotonin, norepinephrine), sphingolipids, aminolevulinic acid...”
When we ran the experiment, we noticed two things:
BioGPT is often not confident in its responses, which means that it outputs “maybe” or two answers (both “yes” and “no”) for about 40% of the provided queries;
The overwhelming majority of provided queries are answered positively when the answer is confident.
Figure 6 shows a confusion matrix for queries with confident answer. We generate the queries set with 1:1 positive to negative ratio. Most of the answers BioGPT-QA provides are positive, which means that the system produces too many false positives and is not usable in the discovery setting.
Challenges in benchmarking for hypothesis generation
Binary interactions. Not every discovery can be represented as a pair of terms, but this is something that most of biomedical graph-based knowledge discovery systems work with. It is a significant limitation of the current approach and a motif discovery is a valid potential direction for future work. Moreover, many databases represent their records as binary interactions [ 42 , 43 , 44 , 45 , 46 ], which can be easily integrated into a link prediction problem.
Directionality. Currently, our choice for pairwise interactions is to omit the directionality information to allow more systems to be evaluated with our framework and cover more pairwise interactions. Directionality is an important component of pairwise interactions, especially when they have types and are formulated in a predication form as a triple: (subject-predicate-object) . Currently, we omit the predicate part and only keep pairs of terms for easier generalization. In many cases, a uni-directional edge \(i\rightarrow j\) does not imply non-existence of \(i\leftarrow j\) . Moreover, in the low-dimensional graph representation construction it is clearly preferable to use undirected edges in our context due to the scarcity of biomedical information. Another caveat is that the tools that detect the logical direction of the predicate in the texts are not perfect [ 47 ]. The information about each particular direction can still be recovered from the underlying cross-referencing citations.
Concept normalization . UMLS is a powerful system combining many biomedical vocabularies together. However, it has certain limitations, such as relatively small number of proteins and chemical compounds. We also observe that many UMLS terms are never covered in the scientific abstracts, even though they exist in the Metathesaurus. This limits the number of obtainable interactions significantly. However, UMLS covers many areas of biomedicine, such as genes, diseases, proteins, chemicals and many others and also provides rich metadata. In addition, NLM provides software for information extraction. There are other vocabularies, which have greater coverage in certain areas (e.g., UniProt ID for proteins or PubChem ID for chemicals), but their seamless integration into a heterogeneous network with literature poses additional challenges that will be gradually addressed in the future work.
We have developed and implemented a comprehensive benchmarking system Dyport for evaluating biomedical hypothesis generation systems. This benchmarking system is advancing the field by providing a structured and systematic approach to assess the efficacy of various hypothesis generation methodologies.
In our pipeline we utilized several curated datasets, which provide a basis in testing the hypothesis generation systems under realistic conditions. The informative discoveries have been integrated into the dynamic graph on top of which we introduced the quantification of discovery importance. This approach allowed us to add a new dimension to the benchmarking process, enabling us to not only assess the accuracy of the hypotheses generated but also their relevance and potential impact in the field of biomedical research. This quantification of discovery importance is a critical step forward, as it aligns the benchmarking process more closely with the practical and applied goals of biomedical research.
We have demonstrated the use case of several graph-based link prediction systems’ verification and concluded that such testing is way more productive than traditional link prediction benchmarks. However, the utility of our benchmarking system extends beyond these examples. We advocate for its widespread adoption to validate the quality of hypothesis generation, aiming to broaden the range of scientific discoveries accessible to the wider research community. Our system is designed to be inclusive, welcoming the addition of more diverse cases.
Future work includes integration of the benchmarking process in the hypothesis system visualization [ 48 ], spreading to other than biomedical areas [ 49 ], integration of novel importance measures, and healthcare benchmarking cases.
Swanson DR. Undiscovered public knowledge. Libr Q. 1986;56(2):103–18.
Article Google Scholar
Swanson DR, Smalheiser NR, Torvik VI. Ranking indirect connections in literature-based discovery: the role of medical subject headings. J Am Soc Inform Sci Technol. 2006;57(11):1427–39.
Article CAS Google Scholar
Peng Y, Bonifield G, Smalheiser N. Gaps within the biomedical literature: Initial characterization and assessment of strategies for discovery. Front Res Metrics Anal. 2017;2:3.
Luo R, Sun L, Xia Y, Qin T, Zhang S, Poon H, Liu T-Y. Biogpt: generative pre-trained transformer for biomedical text generation and mining. Brief Bioinform. 2022;23(6):409.
Sybrandt J, Safro I. Cbag: conditional biomedical abstract generation. PLoS ONE. 2021;16(7):0253905.
Sybrandt J, Shtutman M, Safro I. Moliere: automatic biomedical hypothesis generation system. In: Proceedings of the 23rd ACM SIGKDD. KDD ’17, 2017. pp. 1633–1642. ACM, New York, NY, USA. https://doi.org/10.1145/3097983.3098057 .
Sedler AR, Mitchell CS. Semnet: using local features to navigate the biomedical concept graph. Front Bioeng Biotechnol. 2019;7:156.
Article PubMed PubMed Central Google Scholar
Hristovski D, Peterlin B, Mitchell JA, Humphrey SM. Using literature-based discovery to identify disease candidate genes. Int J Med Inform. 2005;74(2):289–98.
Article PubMed Google Scholar
Gordon MD, Dumais S. Using latent semantic indexing for literature based discovery. J Am Soc Inf Sci. 1998;49(8):674–85.
Sybrandt J, Tyagin I, Shtutman M, Safro I. AGATHA: automatic graph mining and transformer based hypothesis generation approach. In: Proceedings of the 29th ACM international conference on information and knowledge management, 2020;2757–64.
Sourati J, Evans J. Accelerating science with human-aware artificial intelligence. Nat Hum Behav. 2023;7:1682–96.
Chen Y, Argentinis JE, Weber G. IBM Watson: how cognitive computing can be applied to big data challenges in life sciences research. Clin Ther. 2016;38(4):688–701.
Xun G, Jha K, Gopalakrishnan V, Li Y, Zhang A. Generating medical hypotheses based on evolutionary medical concepts. In: 2017 IEEE International conference on data mining (ICDM), pp. 535–44 (2017). https://doi.org/10.1109/ICDM.2017.63 .
Cameron D, Kavuluru R, Rindflesch TC, Sheth AP, Thirunarayan K, Bodenreider O. Context-driven automatic subgraph creation for literature-based discovery. J Biomed Inform. 2015;54:141–57. https://doi.org/10.1016/j.jbi.2015.01.014 .
Sebastian Y, Siew E-G, Orimaye SO. Learning the heterogeneous bibliographic information network for literature-based discovery. Knowl-Based Syst. 2017;115:66–79.
Miranda A, Mehryary F, Luoma J, Pyysalo S, Valencia A, Krallinger M. Overview of drugprot biocreative vii track: quality evaluation and large scale text mining of drug-gene/protein relations. In: Proceedings of the seventh biocreative challenge evaluation workshop, 2021;11–21.
Breit A, Ott S, Agibetov A, Samwald M. OpenBioLink: a benchmarking framework for large-scale biomedical link prediction. Bioinformatics. 2020;36(13):4097–8. https://doi.org/10.1093/bioinformatics/btaa274 .
Article CAS PubMed Google Scholar
Sybrandt J, Shtutman M, Safro I. Large-scale validation of hypothesis generation systems via candidate ranking. In: 2018 IEEE international conference on big data, 2018; 1494–1503. https://doi.org/10.1109/bigdata.2018.8622637 .
Kilicoglu H, Shin D, Fiszman M, Rosemblat G, Rindflesch TC. Semmeddb: a pubmed-scale repository of biomedical semantic predications. Bioinformatics. 2012;28(23):3158–60.
Article CAS PubMed PubMed Central Google Scholar
Fannjiang C, Listgarten J. Is novelty predictable? Cold Spring Harb Perspect Biol. 2024;16: a041469.
Jeon D, Lee J, Ahn J, Lee C. Measuring the novelty of scientific publications: a fastText and local outlier factor approach. J Inform. 2023;17: 101450.
Small H, Tseng H, Patek M. Discovering discoveries: Identifying biomedical discoveries using citation contexts. J Inform. 2017;11:46–62.
Bordes A, Usunier N, Garcia-Duran A, Weston J, Yakhnenko O. Translating embeddings for modeling multi-relational data. In: Advances in neural information processing systems, 2013; 2787–2795.
Bonner S, Barrett IP, Ye C, Swiers R, Engkvist O, Hoyt CT, Hamilton WL. Understanding the performance of knowledge graph embeddings in drug discovery. Artif Intell Life Sci. 2022;2: 100036.
Google Scholar
Bonner S, Barrett IP, Ye C, Swiers R, Engkvist O, Bender A, Hoyt CT, Hamilton WL. A review of biomedical datasets relating to drug discovery: a knowledge graph perspective. Brief Bioinform. 2022;23(6):404.
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015;44(D1):457–62. https://doi.org/10.1093/nar/gkv1070 .
Bodenreider O. The unified medical language system (umls): integrating biomedical terminology. Nucleic Acids Res. 2004;32(suppl_1):267–70.
Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36(6):462–77. https://doi.org/10.1016/j.jbi.2003.11.003 .
Xing R, Luo J, Song T. Biorel: towards large-scale biomedical relation extraction. BMC Bioinform. 2020;21(16):1–13.
Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Adv Neural Inf Process Syst 2013;26.
Aronson AR. Effective mapping of biomedical text to the umls metathesaurus: the metamap program. In: Proceedings of the AMIA symposium, 2001;p. 17.
Welling M, Kipf TN. Semi-supervised classification with graph convolutional networks. In: Journal of international conference on learning representations (ICLR 2017), 2016.
Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks. In: International conference on machine learning, pp. 3319–3328, 2017.
Brandes U. A faster algorithm for betweenness centrality. J Math Sociol. 2001;25(2):163–77.
Aksenova M, Sybrandt J, Cui B, Sikirzhytski V, Ji H, Odhiambo D, Lucius MD, Turner JR, Broude E, Peña E, et al. Inhibition of the dead box rna helicase 3 prevents hiv-1 tat and cocaine-induced neurotoxicity by targeting microglia activation. J Neuroimmune Pharmacol. 2019;1–15.
Tyagin I, Kulshrestha A, Sybrandt J, Matta K, Shtutman M, Safro I. Accelerating covid-19 research with graph mining and transformer-based learning. In: Proceedings of the AAAI conference on artificial intelligence, 2022;36:12673–9.
Grover A, Leskovec J. Node2vec: Scalable feature learning for networks. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. KDD ’16, 2016, pp. 855–864. Association for Computing Machinery, New York. https://doi.org/10.1145/2939672.2939754 .
Costabello L, Bernardi A, Janik A, Pai S, Van CL, McGrath R, McCarthy N, Tabacof P. AmpliGraph: a library for representation learning on knowledge graphs, 2019. https://doi.org/10.5281/zenodo.2595043 .
Sybrandt J, Carrabba A, Herzog A, Safro I. Are abstracts enough for hypothesis generation? In: 2018 IEEE international conference on big data, 2018;1504–1513. https://doi.org/10.1109/bigdata.2018.8621974 .
Liu Z, Roberts RA, Lal-Nag M, Chen X, Huang R, Tong W. Ai-based language models powering drug discovery and development. Drug Discovery Today. 2021;26(11):2593–607.
Jin Q, Dhingra B, Liu Z, Cohen WW, Lu X. Pubmedqa: a dataset for biomedical research question answering, 2019; arXiv preprint arXiv:1909.06146 .
Davis AP, Wiegers TC, Johnson RJ, Sciaky D, Wiegers J, Mattingly CJ. Comparative toxicogenomics database (ctd): update 2023. Nucleic Acids Res. 2022. https://doi.org/10.1093/nar/gkac833 .
Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F. Furlong LI The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2019;48(D1):845–55. https://doi.org/10.1093/nar/gkz1021 .
Ursu O, Holmes J, Knockel J, Bologa CG, Yang JJ, Mathias SL, Nelson SJ, Oprea TI. DrugCentral: online drug compendium. Nucleic Acids Research. 2016;45(D1):932–9. https://doi.org/10.1093/nar/gkw993 .
Calderone A, Castagnoli L, Cesareni G. Mentha: a resource for browsing integrated protein-interaction networks. Nat Methods. 2013;10(8):690–1.
Zeng K, Bodenreider O, Kilbourne J, Nelson SJ. Rxnav: a web service for standard drug information. In: AMIA annual symposium proceedings, 2006; vol. 2006, p. 1156.
Kilicoglu H, Rosemblat G, Fiszman M, Shin D. Broad-coverage biomedical relation extraction with SemRep. BMC Bioinform. 2020;21:1–28.
Tyagin I, Safro I. Interpretable visualization of scientific hypotheses in literature-based discovery. BioCretive Workshop VII; 2021. https://www.biorxiv.org/content/10.1101/2021.10.29.466471v1 .
Marasco D, Tyagin I, Sybrandt J, Spencer JH, Safro I. Literature-based discovery for landscape planning, 2023. arXiv preprint arXiv:2306.02588 .
Rehurek R, Sojka P. Gensim-python framework for vector space modelling. NLP Centre, Faculty of Informatics, Masaryk University, Brno, Czech Republic 2011;3(2).
Fey M, Lenssen JE. Fast graph representation learning with PyTorch Geometric. In: ICLR workshop on representation learning on graphs and manifolds, 2019.
Kokhlikyan, N., Miglani, V., Martin, M., Wang, E., Alsallakh, B., Reynolds, J., Melnikov, A., Kliushkina, N., Araya, C., Yan, S., Reblitz-Richardson, O. Captum: a unified and generic model interpretability library for PyTorch, 2020.
Sollis E, Mosaku A, Abid A, Buniello A, Cerezo M, Gil L, Groza T, Güneş O, Hall P, Hayhurst J, Ibrahim A, Ji Y, John S, Lewis E, MacArthur JL, McMahon A, Osumi-Sutherland D, Panoutsopoulou K, Pendlington Z, Ramachandran S, Stefancsik R, Stewart J, Whetzel P, Wilson R, Hindorff L, Cunningham F, Lambert S, Inouye M, Parkinson H, Harris L. The NHGRI-EBI GWAS catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2022;51(D1):977–85. https://doi.org/10.1093/nar/gkac1010 .
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Research. 2020;49(D1):605–12. https://doi.org/10.1093/nar/gkaa1074 .
Fricke S. Semantic scholar. J Med Lib Assoc: JMLA. 2018;106(1):145.
Download references
Acknowledgements
We would like to thank two anonymous referees whose thoughtful comments helped to improve the paper significantly. This research was supported by NIH award #R01DA054992. The computational experiments were supported in part through the use of DARWIN computing system: DARWIN—A Resource for Computational and Data-intensive Research at the University of Delaware and in the Delaware Region, which is supported by NSF Grant #1919839.
This research was supported by NIH award #R01DA054992. The computational experiments were supported in part through the use of DARWIN computing system: DARWIN—A Resource for Computational and Data-intensive Research at the University of Delaware and in the Delaware Region, which is supported by NSF Grant #1919839.
Author information
Authors and affiliations.
Center for Bioinformatics and Computational Biology, University of Delaware, Newark, DE, 19713, USA
Ilya Tyagin
Department of Computer and Information Sciences, University of Delaware, Newark, DE, 19716, USA
You can also search for this author in PubMed Google Scholar
Contributions
IT processed and analyzed the textual and database data, trained models and implemented the computational pipeline. IS formulated the main idea, supervised the project and provided feedback. Both authors contributed to writing, read and approved the final manuscript.
Corresponding authors
Correspondence to Ilya Tyagin or Ilya Safro .
Ethics declarations
Competing interests.
I declare that the authors have no Conflict of interest as defined by BMC, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Availability of data and materials
The dataset(s), materials and code supporting the conclusions of this article is(are) available in the GitHub repository: https://github.com/IlyaTyagin/Dyport .
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Incorporated technologies
To construct the benchmark, we propose a multi-step pipeline, which requires several key technologies to be used. For the text mining part, we use SemRep [ 28 ] and gensim [ 50 ] implementation of word2vec algorithm. For further stages involving graph learning, we utilize Pytorch Geometric framework and Captum explainability library.
UMLS (Unified Medical Language System) [ 27 ] is one of the fundamental technologies provided by NLM, which consolidates and disseminates essential terminology, taxonomies, and coding norms, along with related materials, such as definitions and semantic types. UMLS is used in the proposed work as a system of concept unique identifiers (CUI) bringing together terms from different vocabularies.
SemRep [ 47 ] is an NLM-developed software, performing extraction of semantic predicates from biomedical texts. It also has the named entity recognition (NER) capabilities (based on MetaMap [ 31 ] backend) and automatically performs entity normalization based on the context.
Word2Vec [ 30 ] is an approach for creating efficient word embeddings. It was proposed in 2013 and is proven to be an excellent technique for generating static (context-independent) latent word representations. The implementation used in this work is based on gensim [ 50 ] library.
Pytorch geometric (PyG) [ 51 ] library is built on top of Pytorch framework focusing on graph geometric learning. It implements a variety of algorithms from published research papers, supports arbitrary-scaled graphs and is well integrated into Pytorch ecosystem. We use PyG to train a graph neural network (GNN) for link prediction problem, which we explain in more detail in methods section.
Captum [ 52 ] package is an extension of Pytorch enabling the explainability of many ML models. It contains attribution methods, such as saliency maps, integrated gradients, Shapley value sampling and others. Captum is supported by PyG library and used in this work to calculate attributions of the proposed GNN.
Appendix B: Incorporated data sources
We review and include a variety of biomedical databases, containing curated connections between different kinds of entities.
KEGG (Kyoto Encyclopedia of Genes and Genomes) [ 26 ] is a collection of resources for understanding principles of work of biological systems (such as cells, organisms or ecosystems) and offering a wide variety of entry points. One of the main components of KEGG is a set of pathway maps, representing molecular interactions as network diagrams.
CTD (The Comparative Toxicogenomics Database) [ 42 ] is a publicly available database focused on collecting the information about environmental exposures effects on human health.
DisGenNET [ 43 ] is a discovery platform covering genes and variants and their connections to human diseases. It integrates data from a list of publicly available databases and repositories and scientific literature.
GWAS (Genome-Wide Association Studies) [ 53 ] is a catalog of human genome-wide association studies, developed by EMBL-EBI and NHGRI. Its aim is to identify and systematize associations of genotypes with phenotypes across human genome.
STRING [ 54 ] is a database aiming to integrate known and predicted protein associations, both physical and functional. It utilizes a network-centric approach and assigns a confidence score for all interactions in the network based on the evidence coming from different sources: text mining, computational predictions and biocurated databases.
DrugCentral [ 44 ] is an online drug information resource aggregating information about active ingredients, indications, pharmacologic action and other related data with respect to FDA, EMA and PMDA-approved drugs.
Mentha [ 45 ] is an evidence-based protein interaction browser (and corresponding database), which takes advantage of International Molecular Exchange (IMEx) consortium. The interactions are curated by experts in compliance with IMEx policies enabling regular weekly updates. Compared to STRING, Mentha is focused on precision over comprehensiveness and excludes any computationally predicted records.
RxNav [ 46 ] is a web-service providing an integrated view on drug information. It contains the information from NLM drug terminology RxNorm, drug classes RxClass and drug-drug interactions collected from ONCHigh and DrugBank sources.
Semantic scholar [ 55 ] is a search engine and research tool for scientific papers developed by the Allen Institute for Artificial Intelligence (AI2). It provides rich metadata about publications which enables us to use Semantic Scholar data for network-based citation analysis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Tyagin, I., Safro, I. Dyport: dynamic importance-based biomedical hypothesis generation benchmarking technique. BMC Bioinformatics 25 , 213 (2024). https://doi.org/10.1186/s12859-024-05812-8
Download citation
Received : 31 January 2024
Accepted : 16 May 2024
Published : 13 June 2024
DOI : https://doi.org/10.1186/s12859-024-05812-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Hypothesis Generation
- Literature-based Discovery
- Link Prediction
- Benchmarking
- Natural Language Processing
BMC Bioinformatics
ISSN: 1471-2105
- General enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
Get Hypothesis Multiple Choice Questions (MCQ Quiz) with answers and detailed solutions. Download these Free Hypothesis MCQ Quiz Pdf and prepare for your upcoming exams Like Banking, SSC, Railway, UPSC, State PSC. ... B. Research hypothesis may be directed at finding out differential effects or relationships.
40 MCQ on Research Methodology. Boost your research methodology knowledge with this comprehensive set of 40 multiple-choice questions (MCQs). Test your understanding of key concepts, study designs, data analysis, and ethical considerations in research. ... Q25. A null hypothesis is (A) when there is no difference between the variables (B) the ...
This set of Probability and Statistics Multiple Choice Questions & Answers (MCQs) focuses on "Testing of Hypothesis". 1. A statement made about a population for testing purpose is called? a) Statistic. b) Hypothesis. c) Level of Significance. d) Test-Statistic. View Answer. 2.
The quiz aimed to sharpen your critical thinking skills and reinforce our grasp on essential concepts in the realm of research. By actively participating in this exercise, we deepened your appreciation for the significance of selecting the right research methods to achieve reliable and meaningful results. 1.
Q 6. Research is A. Searching again and again B. Finding solution to any problem C. Working in a scientific way to search for truth of any problem D. None of the above Q 7. In the process of conducting research 'Formulation of Hypothesis" is followed by A. Statement of Objectives B. Analysis of Data C. Selection of Research Tools
Multiple Choice Quizzes. Try these quizzes to test your understanding. 1. A hypothesis is ______. a wished-for result that the researcher concludes the research with. a complicated set of sentences that pulls variables into proposed complex relationships. a conjecture that is grounded in support background originating from secondary research. 2.
Study with Quizlet and memorise flashcards containing terms like What is a hypothesis? 1. The square root of the sum of squares of two shorter sides of a triangle. 2. A prediction made to test a theory. 3. A set of ideas that drive an area of research. 4. A reliability measurement. 5. A set of related statements that explains a variety of occurrences, What is the difference between interval ...
a) Research refers to a series of systematic activity or activities undertaken to find out the solution to a problem. b) It is a systematic, logical and unbiased process wherein verification of hypotheses, data analysis, interpretation and formation of principles can be done. c) It is an intellectual inquiry or quest towards truth,
MCQ TESTING OF HYPOTHESIS MCQ 13. A statement about a population developed for the purpose of testing is called: (a) Hypothesis (b) Hypothesis testing (c) Level of significance (d) Test-statistic MCQ 13. Any hypothesis which is tested for the purpose of rejection under the assumption that it is true is called: (a) Null hypothesis (b) Alternative hypothesis (c) Statistical hypothesis (d ...
Get Types of Hypotheses Multiple Choice Questions (MCQ Quiz) with answers and detailed solutions. ... In scientific research, a hypothesis is generated from a theory and proposes a relationship between variables that can be tested. Theory: Theories provide a broad explanation for patterns observed in nature, and from these patterns, we can ...
Multiple Choice Quiz. Take the quiz to test your understanding of the key concepts covered in the chapter. Try testing yourself before you read the chapter to see where your strengths and weaknesses are, then test yourself again once you've read the chapter to see how well you've understood. Tip: Click on each link to expand and view the ...
Get Hypothesis testing Multiple Choice Questions (MCQ Quiz) with answers and detailed solutions. Download these Free Hypothesis testing MCQ Quiz Pdf and prepare for your upcoming exams Like Banking, SSC, Railway, UPSC, State PSC.
Skills Practiced. The quiz will help you practice the following skills: Reading comprehension - ensure that you draw the most important information from the related research and null hypotheses ...
Research Hypothesis Multiple Choice Questions (MCQ): Research Hypothesis MCQs with Answers PDF, download App & e-Book to study MSc in psychology courses. Research Hypothesis MCQ PDF: A attribute, presuming different values among different people in different times or places, known as; with Answers for online college courses.
Multiple Choice Questions. Research: A Way of Thinking. The Research Process: A Quick Glance. Reviewing the Literature. Formulating a Research Problem. Identifying Variables. Constructing Hypotheses. The Research Design. Selecting a Study Design.
430+ Research Methodology (RM) Solved MCQs. 108. 40.1k. 18. Take a Test Download as PDF. Hide answers. 1 of 5 Sets. 1.
Multiple Choice Questions on Research Methodology. 1. The method that consists of collection of data through observation and experimentation, formulation and testing of hypothesis is called. 2. Information acquired by experience or experimentation is called as. 3.
Mcqs of Hypothesis (2) - Free download as Word Doc (.doc / .docx), PDF File (.pdf), Text File (.txt) or read online for free. This document contains 20 multiple choice questions about statistical hypotheses. The questions cover key concepts like: 1) Definitions of null hypothesis, alternative hypothesis, and statistical hypothesis. 2) Classifications of hypotheses as simple or composite.
RESEARCH METHODOLOGY MCQ QUESTIONS WITH ANSWERS - Free download as Word Doc (.doc / .docx), PDF File (.pdf), Text File (.txt) or read online for free. N.L. Gage is referred to as "the father of research on teaching". The main purpose of research in education is to help the candidate become an eminent educationist. Inductive inference refers to inferring about the whole population based on the ...
A research hypothesis, in its plural form "hypotheses," is a specific, testable prediction about the anticipated results of a study, established at its outset. It is a key component of the scientific method. Hypotheses connect theory to data and guide the research process towards expanding scientific understanding.
The document contains 22 multiple choice questions about research methodology. It covers topics such as common research methods like surveys, experiments, and longitudinal studies. It also discusses key concepts in research like formulating research questions, hypotheses testing, sampling techniques, and eliminating bias. The questions aim to improve the reader's knowledge of how to properly ...
Research is a process consisting of identifying and defining the research problem, formulating and testing the hypothesis through data collection, organization and analysis, making deductions and reaching of conclusion from the test results of the hypotheses, and reporting and evaluating the research.
Bibliography. A scientific hypothesis is a tentative, testable explanation for a phenomenon in the natural world. It's the initial building block in the scientific method. Many describe it as an ...
Xuepeng Liu The Research Brief. Most manufacturing activities use service inputs such as financial and business services. Dr. Xuepeng Liu's paper [1] examines the implications of services development for the export performance of manufacturing sectors. They develop a methodology to quantify the indirect role of services in international trade ...
Research-Methodology-MCQ - Free download as PDF File (.pdf), Text File (.txt) or read online for free. This document provides multiple choice questions to test understanding of research methodology concepts. It covers topics like the objectives of research, different types of research (applied vs basic, exploratory vs conclusive), formulating research problems and hypotheses, research design ...
Automated hypothesis generation (HG) focuses on uncovering hidden connections within the extensive information that is publicly available. This domain has become increasingly popular, thanks to modern machine learning algorithms. However, the automated evaluation of HG systems is still an open problem, especially on a larger scale. This paper presents a novel benchmarking framework Dyport for ...
385 Mcqs on Research Methodology - Free download as PDF File (.pdf), Text File (.txt) or read online for free. This document contains 25 multiple choice questions about research methodology. The questions cover topics such as the main concepts and objectives of research, different types of research studies and their aims (e.g. descriptive, diagnostic, experimental), characteristics of good ...
A common low-calorie sweetener called xylitol, found in gum, candy, toothpaste and more, may cause clots that can lead to heart attack and stroke, a new study found.