Request for Feedback: Optional object type attributes with defaults in v1.3 alpha
I’m the Product Manager for Terraform Core, and we’re excited to share our v1.3 alpha , which includes the ability to mark object type attributes as optional, as well as set default values ( draft documentation here ). With the delivery of this much requested language feature, we will conclude the existing experiment with an improved design. Below you can find some background information about this language feature, or you can read on to see how to participate in the alpha and provide feedback.
To mark an attribute as optional, use the additional optional(...) modifier around its type declaration:
As many of you know, in Terraform v0.14 we introduced experimental support for an optional modifier for object attributes, which replaced missing attributes with null . Your helpful feedback validated the idea itself, but highlighted a need for specifying defaults for optional attributes. In Terraform v0.15, we added an experimental defaults() function , which allows filling null attribute values with defaults. This resulted in extensive community feedback , and many of you found the defaults function confusing when used with complex structures, or inappropriate for your needs.
We know this experiment has been out in the wild for some time, and we’re incredibly grateful for your patience and feedback on the necessity of this language feature. With that, we’d love for you to try the new syntax available in the v1.3 alpha , and provide any and all feedback.
How to provide feedback
This feature is currently experimental, with the goal of stabilizing a finished feature in the v1.3.0 release. That being said, your feedback and bug reports are vital to us confidently releasing this feature as non-experimental during this release cycle.
Experience reports
Please try out this feature in the alpha release, and let us know what you think. For example:
- Does this new design solve your problems?
- Do you have any feedback on the semantics?
- Is the documentation sufficiently clear?
Bug reports
- For any bugs, please open issues in our repository here .
- For any edge cases that are not solved by this design, you can also open an issue in our repo .
General feedback For general feedback, please comment directly on this post. If you’d prefer to have a private discussion, you can email me directly ( [email protected] ). However, public posts are most helpful for our team to review.
Because this feature is currently experimental, it requires an explicit opt-in on a per-module basis. To use it, write a terraform block with the experiments argument set as follows:
Until the experiment is concluded, the behavior of this feature may see breaking changes even in minor releases. We recommend using this feature only in prerelease versions of modules as long as it remains experimental.
Thank you again for all your contributions to Terraform, and we’re so excited for you to try this release.
I have been using this feature since Aug last year since I couldn’t find a better way to address my use case. I have been subscribed to the PR where this feature was being tracked since then and as soon as I got the notification from you @korinne , I changed the implementation I have before to support the new changes you announced. I love the fact that now I can set a default value much easier than I was doing before . I will be impatiently waiting for this new 1.3.0 release
I have been using this feature in my modules and using coalesce to define default values. This new syntax is much better to work with. Looking forward to its release.
That’s great to hear, thank you!
Hi @korinne ,
We use a lot of nested objects in the module I help maintain. I know this is complex…
In the current implementation, any nested objects have to be defined, even if all of their contained properties are also optional.
In this scenario, I’d like Terraform to infer that, since var.nested_object.settings.setting[1,2] are optional, that I do not need to specify settings key in the default.
Ideally this would work with multiple levels of nested object too.
Is this possible?
If I understand your goals correctly, then yes, this is possible! You can make the settings attribute optional, and specify an empty object {} as its default. Terraform will then apply the specified default attributes inside settings .
Consider this configuration:
Without specifying a variable value:
Only specifying the enabled attribute:
Overriding a settings default:
Does this work with your use case?
@alisdair Just wanted to say thanks again - works a treat!
A quick update on this feature: the most recent alpha build has concluded the experiment, so the terraform { experiments = … } setting should be removed.
Feedback on this change is still very much welcomed during the 1.3 prerelease phase!
@alisdair @korinne Does it work with
For example, Can I do this?
Yes, that’s a valid variable type constraint. You can use optional(<type>, <default>) on any attribute of any object at any level of a type constraint.
I recommend download the latest alpha build and trying it out to check if the behaviour of the defaults is suitable for your use case.
I just pulled in the latest 1.3 Alpha, and made the necessary changes (removed defaults() and replaced with the new concise default declaration syntax, and removed the experiments setting) and I love it!
I’ve been using this feature for a long time now, and being able to set the defaults in my variable declarations, rather than later in locals , makes this code much more easy for my team to read and operate.
Hi, I just found this as I was looking through setting defaults for an object used to define EKS Jobs in a Step Function module. Is it possible to set defaults for nested objects (like two levels in)?
In the above, var.jobs[0].containers.resources still comes out as null. Security context is fine however, resources remains null. See below from terraform console test:
Actually, I think I figured it out and the following configuration works. The feature looks cool. Any idea when 1.3.0 would be ready? Going to add this alpha version to our terraform cloud organization for now
Hi @EmmanuelOgiji ,
It looks like you figured out that you can set a default value for the object as a whole in order to make it appear as a default object when omitted, rather than as null .
An extra note on top of that is to notice that Terraform will automatically convert the default value you specify to the type constraint you specified, and so if your nested object has optional attributes with defaults itself you do not need to duplicate them in the default value of the object, and can instead provide an empty object as the default and have Terraform insert the default attribute values automatically as part of that conversion:
Notice in the above that the default value specified for resources is {} . Terraform will try to convert that to the specified object type, and so in the process it will have the default values of limits and requests inserted into it in the same way that a caller-specified empty object would, and so omitting resources altogether would produce the same final default value as the one you wrote out manually in your example, without the need to duplicate those defaults.
For completeness and for anyone else who is reading who might have a different situation: note that {} was only a valid default value here because all of the attributes in the resources object type are marked as optional. If there were any required attributes in there then the default value would need to include them in order to make the default value convertible to the attribute’s type constraint.
Hi everyone,
I’ve created an issue but it seems my point is more relevant here: Release of optional attributes in TF 1.3 breaks modules using the experimental feature even if compatible · Issue #31355 · hashicorp/terraform · GitHub
TL;DR: IMHO, Terraform 1.3 should introduce a warning instead of an error when experiment is enabled within a module.
Context: we maintain around 70 modules and have activated the experimental feature in a bunch of them lately. We have CI on all of them and on of the test is that module is compatible with latest providers and Terraform version. CI has began to break for latest Terraform aplha version. We fully agreed that the feature was experimental and could break at any time … but we did it despites this, and I think we’re not the only ones.
In Terraform 1.3, having the experiment enabled in a module prevent the user to init its stack, and so, prevent the user to be able to use Terraform 1.3 although the syntax is almost the same (except for managing the default value, which is by the way a lot better in new implementation). My point is that Terraform should display a warning message when encountering the experiment flag instead of breaking with an error since modules using it are mostly compatible.
Hi @BzSpi ,
I replied in the GitHub issue before seeing your feedback here and so I won’t repeat all of what I said over there, but I will restate the most important part: any module using new features introduced in a particular Terraform version will always require using that version, and so as usual a module which uses optional attributes will inherently require using Terraform v1.3 or later because that is the first version that truly supported the feature.
It is interesting that in this particular case there is some overlap between the experimental design and the final design, but experimental features are not part of the language and any module using them should expect to become “broken” either by future iterations of the experiment or by the experiment concluding and that experimental functionality therefore no longer being needed for its intended purpose, which is requesting early feedback in discussion threads such as this one. In recognition of that not having been clear in the past, we are planning to make experiments available only in alpha releases in the future, with stable releases only supporting the stable language.

Terraform block reference
This topic provides reference information about the terraform block. The terraform block allows you to configure Terraform behavior, including the Terraform version, backend, integration with HCP Terraform, and required providers.
Configuration model
The following list outlines attribute hierarchy, data types, and requirements in the terraform block. Click on an attribute for details.
- required_version : string
- required_providers : map
- provider_meta "<LABEL>" : map
- backend "<BACKEND_TYPE>" : map
- organization : string | required when connecting to HCP Terraform
- tags : list of strings
- name : string
- project : string
- hostname : string | app.terraform.io
- token : string
- experiments : list of strings
Specification
This section provides details about the fields you can configure in the terraform block. Specific providers and backends may support additional fields.
Parent block that contains configurations that define Terraform behavior. You can only use constant values in the terraform block. Arguments in the terraform block cannot refer to named objects, such as resources and input variables. Additionally, you cannot use built-in Terraform language functions in the block.
terraform{}.required_version
Specifies which version of the Terraform CLI is allowed to run the configuration. Refer to Version constraints for details about the supported syntax for specifying version constraints.
Use Terraform version constraints in a collaborative environment to ensure that everyone is using a specific Terraform version, or using at least a minimum Terraform version that has behavior expected by the configuration.
Terraform prints an error and exits without taking actions when you use a version of Terraform that does not meet the version constraints to run the configuration.
Modules associated with a configuration may also specify version constraints. You must use a Terraform version that satisfies all version constraints associated with the configuration, including constraints defined in modules, to perform operations. Refer to Modules for additional information about Terraform modules.
The required_version configuration applies only to the version of Terraform CLI and not versions of provider plugins. Refer to Provider Requirements for additional information.
- Data type: String
- Default: Latest version of Terraform
terraform{}.required_providers
Specifies all provider plugins required to create and manage resources specified in the configuration. Each local provider name maps to a source address and a version constraint. Refer to each Terraform provider’s documentation in the public Terraform Registry , or your private registry, for instructions on how to configure attributes in the required_providers block.
- Data type: Map
terraform{}.provider_meta "<LABEL>"
Specifies metadata fields that a provider may expect. Individual modules can populate the metadata fields independently of any provider configuration. Refer to Provider Metadata for additional information.
terraform{}.backend "<BACKEND_TYPE>"
Specifies a mechanism for storing Terraform state files. The backend block takes a backend type as an argument. Refer to Backend Configuration for details about configuring the backend block.
You cannot configure a backend block when the configuration also contains a cloud configuration for storing state data.
terraform{}.cloud
Specifies a set of attributes that allow the Terraform configuration to connect to either HCP Terraform or a Terraform Enterprise installation. HCP Terraform and Terraform Enterprise provide state storage, remote execution, and other benefits. Refer to the HCP Terraform and Terraform Enterprise documentation for additional information.
You can only provide one cloud block per configuration.
You cannot configure a cloud block when the configuration also contains a backend configuration for storing state data.
The cloud block cannot refer to named values, such as input variables, locals, or data source attributes.
terraform{}.cloud{}.organization
Specifies the name of the organization you want to connect to. Instead of hardcoding the organization as a string, you can alternatively use the TF_CLOUD_ORGANIZATION environment variable.
- Required when connecting to HCP Terraform
terraform{}.cloud{}.workspaces
Specifies metadata for matching workspaces in HCP Terraform. Terraform associates the configuration with workspaces managed in HCP Terraform that match the specified tags, name, or project. You can specify the following metadata in the workspaces block:
| Attribute | Description | Data type |
|---|---|---|
| Specifies a list of flat single-value tags. Terraform associates the configuration with workspaces that have all matching flat single-value tags. New workspaces created from the working directory inherit the tags. This attribute does not support key-value tags. You cannot set this attribute and the attribute in the same configuration. | Array of strings | |
| Specifies an HCP Terraform workspace name to associate the Terraform configuration with. You can only use the working directory with the workspace named in the configuration. You cannot manage the workspace from the Terraform CLI. You cannot set this attribute and the attribute in the same configuration. Instead of hardcoding a single workspace as a string, you can alternatively use the environment variable. | String | |
| Specifies the name of an HCP Terraform project. Terraform creates all workspaces that use this configuration in the project. Using the command in the working directory returns only workspaces in the specified project. Instead of hardcoding the project as a string, you can alternatively use the environment variable. | String |
terraform{}.cloud{}.hostname
Specifies the hostname for a Terraform Enterprise deployment. Instead of hardcoding the hostname of the Terraform Enterprise deployment, you can alternatively use the TF_CLOUD_HOSTNAME environment variable.
- Required when connecting to Terraform Enterprise
- Default: app.terraform.io
terraform{}.cloud{}.token
Specifies a token for authenticating with HCP Terraform. We recommend omitting the token from the configuration and either using the terraform login command or manually configuring credentials in the CLI configuration file instead.
terraform{}.experiments
Specifies a list of experimental feature names that you want to opt into. In releases where experimental features are available, you can enable them on a per-module basis.
Experiments are subject to arbitrary changes in later releases and, depending on the outcome of the experiment, may change significantly before final release or may not be released in stable form at all. Breaking changes may appear in minor and patch releases. We do not recommend using experimental features in Terraform modules intended for production.
Modules with experiments enabled generate a warning on every terraform plan or terraform apply operation. If you want to try experimental features in a shared module, we recommend enabling the experiment only in alpha or beta releases of the module.
Refer to the Terraform changelog for information about experiments and to monitor the release notes about experiment keywords that may be available.
- Data type: List of strings
Environment variables for the cloud block
You can use environment variables to configure one or more cloud block attributes. This is helpful when you want to use the same Terraform configuration in different HCP Terraform organizations and projects. Terraform only uses these variables if you do not define corresponding attributes in your configuration. If you choose to configure the cloud block entirely through environment variables, you must still add an empty cloud block in your configuration file.
You can use environment variables to automate Terraform operations, which has specific security considerations. Refer to Non-Interactive Workflows for details.
Use the following environment variables to configure the cloud block:
TF_CLOUD_ORGANIZATION - The name of the organization. Terraform reads this variable when organization is omitted from the cloud block ` . If both are specified, the configuration takes precedence.
TF_CLOUD_HOSTNAME - The hostname of a Terraform Enterprise installation. Terraform reads this when hostname is omitted from the cloud block. If both are specified, the configuration takes precedence.
TF_CLOUD_PROJECT - The name of an HCP Terraform project. Terraform reads this when workspaces.project is omitted from the cloud block. If both are specified, the cloud block configuration takes precedence.
TF_WORKSPACE - The name of a single HCP Terraform workspace. Terraform reads this when workspaces is omitted from the cloud block. HCP Terraform will not create a new workspace from this variable; the workspace must exist in the specified organization. You can set TF_WORKSPACE if the cloud block uses tags. However, the value of TF_WORKSPACE must be included in the set of tags. This variable also selects the workspace in your local environment. Refer to TF_WORKSPACE for details.
The following examples demonstrate common configuration patterns for specific use cases.
Add a provider
The following configuration requires the aws provider version 2.7.0 or later from the public Terraform registry:
Connect to HCP Terraform
In the following example, the configuration links the working directory to workspaces in the example_corp organization that contain the app tag:
Connect to Terraform Enterprise using environment variables
In the following example, Terraform checks the TF_CLOUD_ORGANIZATION and TF_CLOUD_HOSTNAME environment variables and automatically populates the organization and hostname arguments. During initialization, the local Terraform CLI connects the working directory to Terraform Enterprise using those values. As a result, Terraform links the configuration to either HCP Terraform or Terraform Enterprise and allows teams to reuse the configuration in different continuous integration pipelines:
Flavius Dinu - Tech Blog
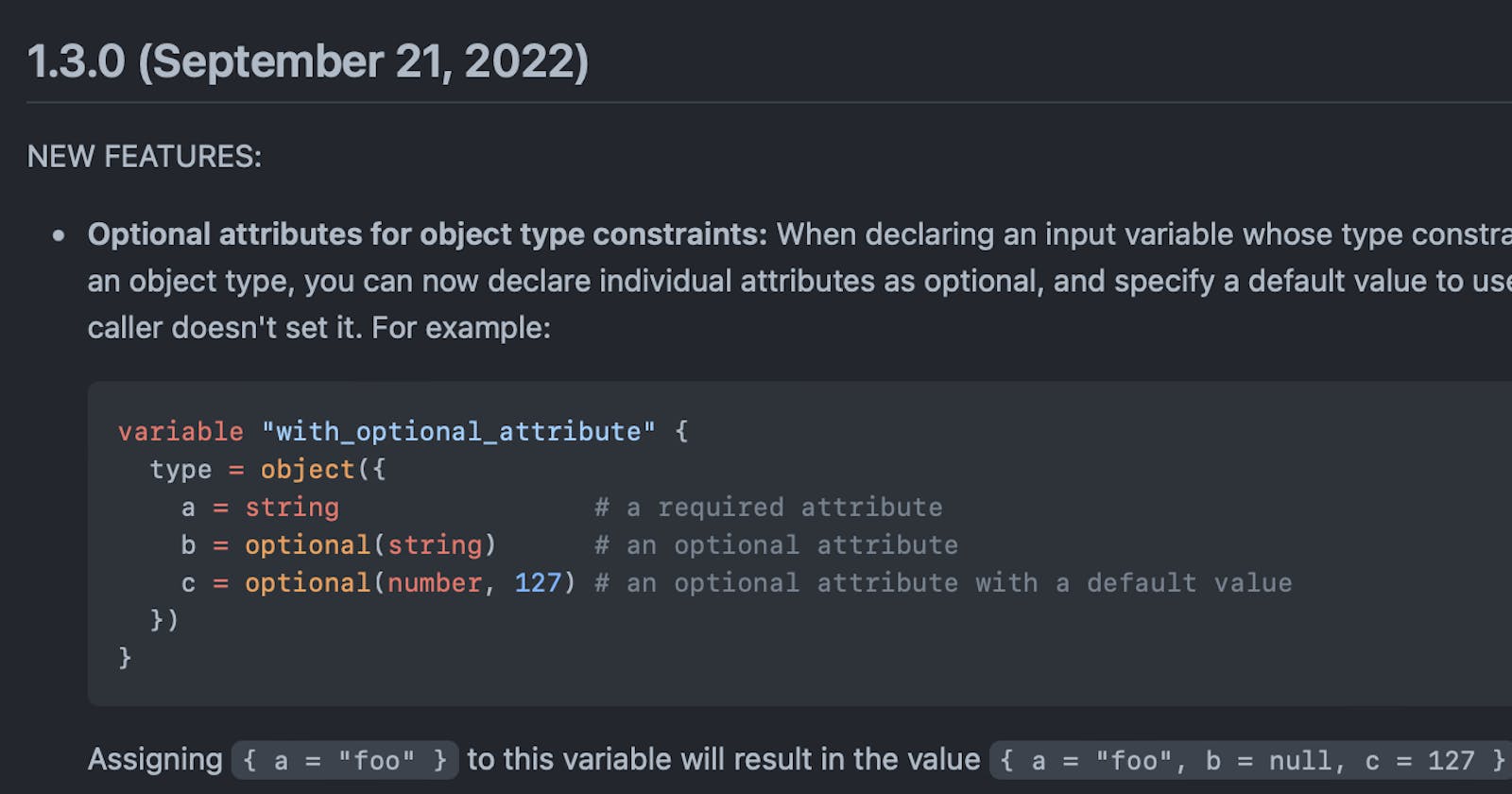
Terraform Optional Object Type Attributes

Table of contents
What does this actually do, why is this important, old main.tf, new main.tf, old variables.tf, new variables.tf, useful links.
If you read my previous posts, you already know that I am big fan of using for_each and object variables when I'm building my modules.
For quite some time, I've been waiting for a particular feature to exit beta and this is the optional object type attribute. The Optional Object type attribute, was in beta for quite some time since Terraform 0.14 and now in Terraform 1.3 (released end of September 2022) it's GA. So to get this straight, this is not a new feature, but now it is 100% ready to be used in production use cases.
When you are building a generic module and you want to offer a lot of possibilities for the people that are going to use it, you will use objects.
Nevertheless, this created a big problem in the past: all the attributes had to be provided by the person using that module and of course, no one will ever need to configure everything a module offers. This meant that you had to use an any type, but if you like to generate documentation with tfdocs , the variables part wouldn't be very helpful. The module code was also pretty ugly, with a lot of lookups to set default values and whatnot.
There are two things that you can actually do with this feature when you are using object variables:
- Set object parameter as optional
- Set default values to the object's parameters
You can build better modules, with less code and the documentation will be astonishing when you generate it with tfdocs, making you aware of all of the configurable parameters.
Example Usage
I will show you how I've changed a Terraform Module and what are the differences on the main.tf and variables.tf files.
As you see, in the old version of the code, I had a lot of lookups and I had to provide the default values on the main.tf version.
0 lookups, less code, easier to read and understand.
In the above variables.tf , I've provided the parameters to the description of the variable, but of course, I did this to help users understand the attributes of my variable. This is not how it's supposed to be done, but I wanted to make tfdocs generate a somewhat readable documentation.
In the new example, we are using the powerful optional attribute. The same thing that was done by a lookup in the resource code, we can now do with this keyword. This is the Optional syntax: optional(parameter_type, default_value) . The simplicity of it, is exactly what we needed in order to speed up module development and to keep up with the maintenance for them.
I am very thrilled with this feature and I totally recommend checking it out.
- Example Helm Release Module Code
- Terraform v1.3.0 Release Info
- Type Constraints
Fix “Cannot use import statement outside a module” Error (2024 Guide)
- Itamar Haim
Have you ever seen this error message while coding in JavaScript? "Cannot use import statement outside a module." This error often pops up when you're trying to use modules in your JavaScript code. It can stop your work and leave you confused. No worries! This easy-to-follow guide will help you figure out what's causing this error and how to fix it.

Understanding JavaScript Modules
JavaScript modules are like building blocks for your code . They help you organize your work and reuse parts of it easily. In JavaScript, modules come in two main flavors:
- ES6 Modules : These are the newer, more modern type.
- CommonJS : This is an older type that is still used in many projects.
The error we’re talking about usually happens when these two types clash.
ES6 Modules: The New Way
ES6 modules came with ECMAScript 6 (also called ES2015). They offer a clean way to share code between files. Here’s what makes them great:
- Better Code Organization : You can split your code into smaller, easier-to-manage pieces.
- Easy Reuse : You can use the same code in different parts of your project or even in new projects.
- Clear Dependencies : It’s easy to see which parts of your code depend on others.
Here’s a quick example of ES6 modules:
In this example, math.js is a module that shares two functions. app.js then uses these functions.
CommonJS: The Old Reliable
CommonJS has been around longer, especially in Node.js. It uses different keywords:
- require to bring in code from other files
- module.exports to share code with other files
Here’s how it looks:
In this case, utils.js shares a greet function, which app.js then uses.
Key Differences Between ES6 and CommonJS
Understanding these differences can help you avoid the “Cannot use import statement outside a module” error:
- Use import and export
- Load code at compile time
- Work in browsers with <script type=”module”>
- Need some setup to work in Node.js
- Great for new projects and big apps
- Use require and module.exports
- Load code at runtime
- Work in Node.js out of the box
- Need extra tools to work in browsers
- Good for existing Node.js projects and simple scripts
Choosing the Right Module System
When starting a new project:
- Use ES6 modules if you have a specific reason not to.
For an existing Node.js project:
- If it’s already using CommonJS and is simple enough, stick with CommonJS .
For browser scripts:
- Use ES6 modules with <script type=”module”> or a module bundler.
Try to use just one system in your project to keep things simple.
Fixing the Error in Node.js
Node.js now supports both CommonJS and ES6 modules. This can sometimes cause the “Cannot use import statement outside a module” error. So, you’re trying to use the import feature, which is part of ES6, in a file that Node.js thinks is using CommonJS. That’s what causes this error.
To fix this, you need to tell Node.js which module system you’re using. We’ll cover how to do that in the next section.
How to Fix the “Cannot use import statement outside a module” Error
Let’s look at three ways to fix this common JavaScript error. Each method has its own pros and cons, so choose the one that fits your needs best.
Solution 1: Use ES6 Modules in Node.js
The easiest way to fix this error is to tell Node.js that you’re using ES6 modules. Here’s how:
- Open your package.json file.
- Add this line:
This tells Node.js to treat all .js files as ES6 modules. Now, you can use import and export without errors.
Tip : If you need to mix ES6 and CommonJS modules, use these file extensions:
- .mjs for ES6 modules
- .cjs for CommonJS modules
Solution 2: Use the –experimental-modules Flag
If you’re rocking an older version of Node.js (before 13.2.0), don’t fret! You can still take advantage of ES6 modules. Just add a flag when you run your code:
This flag tells Node.js to treat .mjs files as ES6 modules.
Important notes:
- This flag might not work the same as newer Node.js versions.
- It might not be available in future Node.js versions.
When to use this flag:
- You’re working on an old project with an older Node.js version.
- You want to test the ES6 module code quickly.
- You’re learning about ES6 modules in an older Node.js setup.
Solution 3: Use Babel to Convert Your Code
Sometimes, you can’t update Node.js or use experimental flags. You may be working on an old project, or some of your code only works with an older version. In these cases, you can use a tool called Babel.
Babel changes your modern JavaScript code into older code that works everywhere. Here’s what it does:
Your code now works in older Node.js versions without the “Cannot use import statement outside a module” error.
How to set up Babel:
- Install Babel packages.
- Create a Babel config file ( .babelrc or babel.config.js ).
- Add settings to change ES6 modules to CommonJS.
Things to think about:
- Using Babel adds an extra step when you build your project.
- Your code might run slower, but you won’t notice.
When to use Babel:
- You’re working on an old Node.js project you can’t update.
- Some of your code only works with an older Node.js version.
- You want to write modern JavaScript but need it to work in older setups.
How to Fix Module Errors in Web Browsers
Modern web browsers can use ES6 modules, but you need to set things up correctly. Let’s look at how to fix the “Cannot use import statement outside a module” error in your web projects.
New web browsers support ES6 modules, but you need to tell the browser when you’re using them. You do this with a special script tag. This tag lets the browser load modules, handle dependencies, and manage scopes the right way.
Solution 1: Use the <script type=”module”> Tag
The easiest way to use ES6 modules in a browser is with the <script type=”module”> tag. Just add this to your HTML:
This tells the browser, “This script is a module.” Now you can use import and export in my_script.js without getting an error.
Here’s an example:
In this example, utils.js shares the greet function, and my_script.js uses it. The <script type=”module”> tag makes sure the browser knows my_script.js is a module.
Important things to know:
- Script Order: When you use multiple <script type=”module”> tags, the browser runs them in the order they appear in the HTML. This ensures that everything loads in the right order.
- CORS: If you load modules from a different website, that website needs to allow it. This is called Cross-Origin Resource Sharing (CORS).
The <script type=”module”> tag works well for small projects or when you want to load modules directly. For bigger projects with lots of modules, use a module bundler.
Solution 2: Use Module Bundlers
As your web project grows and has many modules that depend on each other, it can take effort to manage all the script tags. This is where module bundlers come in handy.
What Are Module Bundlers?
Module bundlers are tools that examine all the modules in your project, determine how they connect, and pack them into one or a few files. They also handle loading and running modules in the browser. Some popular bundlers are Webpack, Parcel, and Rollup.
How Bundlers Help
- They Figure Out Dependencies: Bundlers make sure your modules load in the right order, even if they depend on each other in complex ways.
- They Make Your Code Better: Bundlers can make your files smaller and faster to load.
- They Make Your Code Work Everywhere: Bundlers can change your code to work in older browsers that don’t support ES6 modules.
Choosing a Bundler
Different bundlers are good for different things:
- Webpack: Good for big, complex projects. You can change a lot of settings.
- Parcel: Easy to use. You don’t have to set up much.
- Rollup: Makes small, efficient code. Often used for making libraries.
Using Bundlers with Elementor
If you’re using Elementor to build a WordPress website, you can still use module bundlers. Elementor works well with bundlers to make sure your JavaScript code loads quickly and efficiently.
JavaScript Modules: Best Practices and Troubleshooting
Even if you understand module systems, you might still run into problems. Let’s look at some common issues that can cause the “Cannot use import statement outside a module” error and how to fix them. We’ll also cover good ways to organize your code with modules.
Common Problems and Solutions
Here are some typical issues that can lead to the “Cannot use import statement outside a module” error:
- Problem: Using import in a CommonJS module or require in an ES6 module.
- Solution: Pick one system and stick to it. If you must mix them, use tools like Babel to make your code work everywhere.
- Problem: Using the wrong extension for your module type in Node.js.
- Solution: If you haven’t set “type”: “module” in your package.json, use .mjs for ES6 modules and .cjs for CommonJS modules.
- Problem: Forgetting to set up your project correctly for modules.
- Solution: Check your package.json file for the right “type” setting. Also, make sure your bundler settings are correct if you’re using one.
- Problem: Modules that depend on each other in a loop.
- Solution: Reorganize your code to break the loop. You should create a new module for shared code.
Organizing Your Code with Modules
Modules aren’t just for fixing errors. They help you write better, cleaner code. Here are some tips:
- Good: stringUtils.js , apiHelpers.js
- Not so good: utils.js , helpers.js
- Group related modules together.
- You could organize by feature, function, or layer (like components, services, utilities).
- Each module should do one thing well.
- If a module gets too big, split it into smaller ones.
- Don’t let modules depend on each other in a loop.
- If you need to, create a new module for shared code.
- Clearly show what each module shares and uses.
- Try to only use import * from … unless you really need to.
The Future of JavaScript Modules
ES6 modules are becoming the main way to use modules in JavaScript. They work in most browsers now and are getting better support in Node.js. Here’s why they’re good:
- They have a clean, easy-to-read syntax.
- They load modules in a way that’s easier for computers to understand.
- They clearly show what each module needs.
If you’re starting a new project, use ES6 modules. If you’re working on an old project that uses CommonJS, think about slowly changing to ES6 modules. Tools like Babel can help with this change.
Elementor: Making Web Development Easier
If you want to build websites faster and easier, you might like Elementor . It’s a tool that lets you design websites without writing code. But it’s not just for design – it also helps with technical stuff like JavaScript modules.
H ow Elementor Simplifies Module Management
Elementor streamlines module handling, taking care of much of the loading and interaction behind the scenes, especially when using its built-in elements and features. This simplifies development and reduces the chance of encountering common module-related issues.
Elementor AI: Your Development Assistant
Elementor also provides AI capabilities to speed up your workflow:
- Code Suggestions: Get help writing code for elements like animations.
- Content Help: Generate text for your website.
- Design Ideas: Receive suggestions for layouts and color schemes.
These AI features can boost productivity and inspire new ideas.
Remember: While Elementor simplifies module management, certain errors may still arise with custom JavaScript or external libraries . Additionally, AI assistance is valuable but may require human review and refinement.
Overall, Elementor’s combination of module handling and AI features empowers developers and designers to build websites more efficiently and creatively.
We’ve covered a lot about the “Cannot use import statement outside a module” error. We looked at why it happens and how to fix it in Node.js and in browsers. We also talked about good ways to use modules in your code.
Remember, ES6 modules are becoming the main way to use modules in JavaScript . They’re cleaner and more future-proof if you can start using them in your projects.
If you want to make building websites easier, check out Elementor. It can help with both design and technical stuff, like modules.
Keep learning and practicing, and you’ll get better at handling modules and building great websites!
Looking for fresh content?
By entering your email, you agree to receive Elementor emails, including marketing emails, and agree to our Terms & Conditions and Privacy Policy .
- Incredibly Fast Store
- Sales Optimization
- Enterprise-Grade Security
- 24/7 Expert Service
- Unlimited Websites
- Unlimited Upload Size
- Bulk Optimization
- WebP Conversion

- Prompt your Code & Add Custom Code, HTML, or CSS with ease
- Generate or edit with AI for Tailored Images
- Use Copilot for predictive stylized container layouts
- Craft or Translate Content at Lightning Speed
- Super-Fast Websites
- Any Site, Every Business
- Drag & Drop Website Builder, No Code Required
- Over 100 Widgets, for Every Purpose
- Professional Design Features for Pixel Perfect Design
- Marketing & eCommerce Features to Increase Conversion
Don't Miss Out!
Subscribe to get exclusive deals & news before anyone else!
Thank You! You’re In!
Don’t just sit back—dive right into our current plans now!
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 12 September 2024

Inferring gene regulatory networks with graph convolutional network based on causal feature reconstruction
- Ruirui Ji 1 , 2 ,
- Yi Geng 1 &
- Xin Quan 1
Scientific Reports volume 14 , Article number: 21342 ( 2024 ) Cite this article
1 Altmetric
Metrics details
- Computational biology and bioinformatics
- Data mining
- Gene regulatory networks
- Machine learning
- Systems biology
Inferring gene regulatory networks through deep learning and causal inference methods is a crucial task in the field of computational biology and bioinformatics. This study presents a novel approach that uses a Graph Convolutional Network (GCN) guided by causal information to infer Gene Regulatory Networks (GRN). The transfer entropy and reconstruction layer are utilized to achieve causal feature reconstruction, mitigating the information loss problem caused by multiple rounds of neighbor aggregation in GCN, resulting in a causal and integrated representation of node features. Separable features are extracted from gene expression data by the Gaussian-kernel Autoencoder to improve computational efficiency. Experimental results on the DREAM5 and the mDC dataset demonstrate that our method exhibits superior performance compared to existing algorithms, as indicated by the higher values of the AUPRC metrics. Furthermore, the incorporation of causal feature reconstruction enhances the inferred GRN, rendering them more reasonable, accurate, and reliable.
Similar content being viewed by others

Integration of multiomics data with graph convolutional networks to identify new cancer genes and their associated molecular mechanisms
CGMega: explainable graph neural network framework with attention mechanisms for cancer gene module dissection
Deep learning of causal structures in high dimensions under data limitations
Introduction.
Gene regulatory network (GRN) 1 describe the complex regulatory relationships among genes and is one of the key tools to assist researchers in analyzing and understanding biological processes at the molecular level. The advancement of high-throughput sequencing technology has resulted in the accumulation of a substantial volume of gene expression data. Mining the regulatory relationships among genes accurately based on gene expression data has become the research focus of computational biology and bioinformatics. This research is of great significance in promoting the development of biomedicine and uncovering potential biological processes.
At the outset, researchers utilized statistical-based methods 2 to infer GRN. However, purely statistical methods only consider the statistical patterns existing in gene expression data 3 , disregarding the causal relationships among gene expression data which leads to low accuracy and no biological significance in the inference results. Therefore, researchers have begun to focus on analyzing the causal regulatory relationships between genes 4 . For example, Ma et al. 5 proposed a nonlinear differential model based on time series data. This model achieves network inference by establishing a functional relationship between target genes and their regulatory genes. The model parameters are subsequently optimized using the Random Forest algorithm. Friedman et al. 6 used Bayesian networks to establish a causal skeleton and realized network inference based on the network skeleton. Ajmal et al. 7 utilized a dynamic Bayesian network to infer the network and achieve the simulation of somatic regulatory relationships between genes with multiple time lags, however, leads to a significant demand for computational resources as the number of time points increases. Olsen et al. 8 proposed a method for inferring causal edges in a network by analyzing whether a variable is causally influenced by two or more variables based on a network skeleton. Feng et al. 9 inferred the network skeleton using multiple-time Transfer Entropy for each pair of genes and filtered out low-confidence edges in the network using a threshold, and then retained directed edges based on the threshold through enumeration or searching. Sun et al. 10 , 11 proposed the concept of causal entropy and an inference method based on optimal causal entropy, which incorporates time-series data to enhance the perception of causal relationships between variables.
The accuracy of network inference is improved and biologically meaningful results are generated by causal network inference methods. The causal network inference approaches consists of two steps. Firstly, the causal skeleton or the major threshold is determined by the approaches. Secondly, an enumeration or heuristic algorithm identifies additional causal edges based on the skeleton. However, these approaches suffer from two main problems: excessive computational time and resource consumption, and a lack of effective constraints during the inference process, leading to the potential generation of erroneous results.
With the advancement of deep learning, researchers have started using deep learning methods to infer GRN more effectively 12 . Compared to traditional causal network inference methods, deep learning methods can learn more intricate regulatory relationships from expression data with higher accuracy. Wei Liu et al. 13 proposed a circRNA disease-association prediction model based on automatically selected meta-path and contrastive learning, in which GNN is used to extract node features. Li et al. 14 proposed a gated convolutional recurrent network with residual learning to predict translation initiation sites. Guo et al. 15 proposed a variational gated autoencoder-based feature extraction mode to potential disease-miRNA associations. Meroua et al. 16 constructed deep neural networks based on known regulatory pairs for network inference. MacLean et al. 17 extracted regulatory features by regulatory pairs of microarray gene expressions and constructed convolutional neural networks (CNN) for network inference. These methods only use one-to-one known regulatory pairs as labels to construct neural networks for predicting potential regulatory relationships, however, it is challenging to learn the intricate regulatory relationships of genes within the network topology. Graph Neural Networks (GNN) 18 is the network model capable of processing graph data, which enables efficient representation and inference of graph structures. Therefore, the use of graph neural networks to mine more complex regulatory topology based on known gene regulatory relationships is gradually emerging as a new tool for inferring GRN. Wang et al. 19 proposed a GNN link prediction method to predict regulatory relationships between genes using gene expression data as a feature matrix, and employed the network skeleton to calculate the Neighbor Aggregation of each order of genes in the network to perform inference in a semi-supervised approach, which brings in additional data requirements. Chen et al. 20 proposed a graph attention network to infer latent interactions between transcription factors and target genes in GRN, however, graph-based attention mechanisms incur huge computational and memory overheads.Graph Convolutional Neural Networks (GCN) 21 is a graph-based method build on GNN. By convolutional operations and hierarchical aggregation, GCN is more stable and accurate than GNN in generating neighbourhood aggregation, making it widely used in biological networks.. S. Ganeshamoorthy et al. 22 used a 1D-CNN to extract key features from gene expression data. then utilized the extracted features and known regulatory pairs as input for a Graph Variational Autoencoder (GVAE) which is composed of GCN to achieve GRN inference. Mao et al. 23 proposed a GCN-based interaction encoder infer GRN, by neighbor aggregation to capturing interdependencies between nodes in the network, due to which the performance of the model is impacted by the precision of neighbour aggregation. The accuracy of inferring GRN can be enhanced by using graph neural networks to generate a graph representation of known regulatory relationships through neighbor aggregation. However, the neighbor information is easily lost during aggregation and which lead to unreliable accuracy in downstream tasks. Therefore, it is the key to infer GRN by graph neural networks that ensuring the causal relationships are not overlooked during neighbor nodes aggregation, which embedded in known regulatory pairings and expression data.
Transfer Entropy (TE) 24 is a kind of index that quantitatively compute the flow of information from one series to another. Currently, TE has been combined with graph model for prediction tasks. Duan et al. 25 used TE to extract the causality among the time series and construct the TE graph as a priori-information to guide the forecasting task. Zhang et al. 26 proposed a rutting prediction model based on multi-variate transfer entropy and GNN. TE has been used as a pre-processing before graph networks in approaches described above, inspired by which, TE can be introduced to measure the flow of neighbour information during the order-by-order node aggregation in GCN and enhance the accuracy of which. Since Transfer Entropy reflects the direction and strength of the transferred neighbour information at the same time, it indicates the essential causal relationship between neighbour aggregation of each order in GRN. Reinforcing the causal relationship in the neighbour aggregation with a method guided by transfer entropy will render the inferred GRN more reasonable and reliable.
Therefore, in this paper, the GCN based on Causal Feature Reconstruction method is firstly proposed for inferring GRN. The method employs Transfer Entropy to quantify the loss of causal information in the GCN during neighbor aggregation. Subsequently, the representations of the node features are obtained through Causal Feature Reconstruction. This method results in a more comprehensive node feature representation output from the GCN, enhancing the accuracy of the downstream link prediction task.
The main contributions of this paper are as follows:
Firstly proposing a causal information-guided GCN method for inferring GRN by using Transfer Entropy to measure and enhance the neighbor aggregation. It also incorporates linear layers to complete the causal feature reconstruction of neighbor aggregation, aiming to reduce the loss of neighbor information in the GCN during the training process.
Incorporating a Gaussian kernel into an Autoencoder method to extract features from gene expression data. The Gaussian-kernel Autoencoder extracts gene expression data into significantly separable features, which is reliable and comprehensive. The method aims to enhance the subsequent computational efficiency of GCN and precision of causal reconstruction.
Validating the method on the E.coli , the S.cerevisiae , and the mDC datasets, the experimental results demonstrate that the model in this paper achieves higher network inference accuracy and credibility.
Link prediction and inference of GRN using GCN based on causal feature reconstruction
Overall framework.
In this paper, the GCN based on causal feature reconstruction is investigated to infer GRN. The proposed model consists of three main components: a Gaussian-kernel Autoencoder module, a GCN based on causal feature reconstruction, and a link prediction module. The framework of the model is illustrated in Fig. 1 . Firstly, the Gaussian-kernel autoencoder is used to extract the gene expression features \({\textbf{X}}\) from the gene expression data. Subsequently, the gene expression features \({\textbf{X}}\) and the neighbor matrix \({\textbf{A}}\) which is obtained from the known regulatory pairs, are fed into the GCN module. The output of the GCN is reconstructed by causal features, thereafter the inference of the GRN is completed using link prediction.
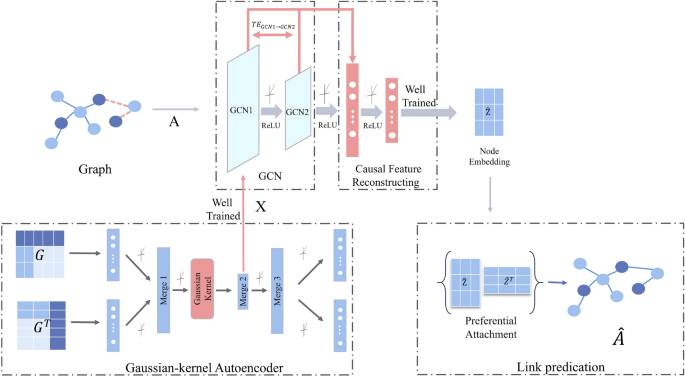
The framework of inferring GRN by GCN based on causal feature reconstruction (where \({\textbf{A}}\) is the neighbourhood matrix extracted from regulatory pairs, \({\textbf{X}}\) is the gene expression feature extracted by the Gaussian-kernel Autoencoder, \({\textbf{G}}\) is the neighbourhood matrix of the inference network, and \({\textbf{G}}^\top\) is the transposition of the gene expression data, \(\hat{\textbf{A}}\) is the inferred network).
Extracting gene expression features
Gene expression features.
Gene expression data describes the intensity of gene expression at a specific condition, with higher values indicating greater intensity. The expression intensity provides insight into the regulatory relationship to some extent. In static gene expression data, the expression values of different genes in the same sample can reflect the activated or inhibited state of genes at a certain condition. This, in turn, reflects a specific regulatory state. Additionally, the expression values of the same genes in different groups of data also vary, indicating the expression state of genes in different states of the GRN 27 . Therefore, extracting key effective features from gene expression data is crucial for accurately inferring the GRN.
Mirzal et al. 28 used non-negative matrix decomposition to process gene expression data, providing guidelines for key gene screening. Fan et al. 29 improved the accuracy of large-scale gene regulatory network inference by performing singular value decomposition on gene expression data. Ganeshamoorthy et al. 22 extracted the key features of gene expression using 1D-CNN and utilized these features as inputs to the GVAE, which led to improved accuracy of the inferred network. The Gaussian kernel 30 enable the data to be linearly separable, which have been used to extract separable feature in multi-omics data integration task 31 . The methods mentioned above are capable of extracting the required expression features. However, it is difficult for the inference algorithm to prioritise the most important features as there are small but significant differences in these features, which in turn affects the accuracy of the inference. The autoencoder has been widely used in Biodata feature extraction, such as a autoencoder-based model to classify the glioma subtype 32 , and a stacked autoencoder to predict the potential miRNA-disease associations 33 , the autoencoder is able to extract deep feature from Biodata.
Therefore, in this paper, the Gaussian kernel is incorporated into the Autoencoder, and the features are enhanced by the Gaussian kernel. This results in the original features becoming distinguishable in a separable manner. The Gaussian-kernel Autoencoder is capable of extracting deep features from expression data in both rows and columns, allowing for differentiation between them. After completing the feature extraction for gene expression, the resulting feature matrix is then used as input for the GCN. This enhances the accuracy and confidence of link prediction.
Gaussian-kernel autoencoder
The structure of the Gaussian-kernel Autoencoder is shown in Fig. 2 . The Autoencoder consists of two inputs, multiple encoders and decoders, and a Gaussian kernel module. The row values of the gene expression data reflect reflect the state of specific gene under different regulatory relationships, however, it is difficult to reflect the state of the gene under different regulatory relationships. Transposing the gene expression data to invert the rows and columns, which reflecting different genes under specific regulatory relationship, thus the gene expression data and its transposition are used as inputs to the encoder to extract the regulatory state features embedded in the row and column values, which are further encoded and fused in depth by the merging layer to obtain more accurate regulatory features. Multiple merge layers share the weights in the encoder 34 . The merge layer consists of MLP layers which fuse and amplify the input before splitting it into output vectors. Next, a Gaussian kernel module is used to capture differential features and ultimately extract key expression features of genes.
The Gaussian-kernel as shown in Eq. ( 1 ):
where \({\sigma }\) is is the parameter of the Gaussian kernel.

The Structure of Gaussian-kernel Autoencoder (where \({\textbf{G}}\) represents gene expression data, \({\textbf{G}}^\top\) represents gene expression data transposition).
The loss function measures the error between the input and output layers of the Gaussian-kernel Autoencoder, which is composed of Mean Squared Error (MSE) and Kullback-Leibler (KL) divergence. When the loss is extremely small, it indicates that the expression features accurately reflect the deep features contained in the rows and columns, as shown in Eq. ( 2 ):
where \(x_{1}\) and \(x_{2}\) denote the input features of the Autoencoder, \(x_1^{\prime }\) and \(x_2^{\prime }\) denote the inferred outputs, W denotes the weight and \(\lambda\) denotes the regularization factor.
GCN module based on causal feature reconstruction

The framework of GCN link prediction based on causal feature reconstruction (where \({\textbf{X}}\) represents the feature matrix, \({\textbf{A}}\) represents the adjacency matrix and \(\hat{\textbf{A}}\) represents the adjacency matrix of the inferred network).
The convolution layer of a GCN calculates the interactions between each node and its neighboring nodes through neighbor aggregation. It then combines this information with the input features to generate a new representation for each node. Multiple layers of neighbor aggregation enable the GCN to progressively gather neighbor aggregation of nodes of all orders. As a result, a more comprehensive feature representation is computed. It is essential that ensuring the validity and causality of the neighbor information to improve the accuracy of link prediction. For this purpose, Transfer Entropy is utilized to quantify the causality of each order of neighbor information aggregation, which allows for a more comprehensive and accurate feature representation of the nodes in the causal reconstruction module. Consequently, it enhances the causality and dependency among the neighbors of each order, ultimately improving the accuracy of GRN inference. The framework of the GCN based on causal feature reconstruction is depicted in Fig. 3 , which includes a GCN module and a causal feature reconstruction module.
Graph convolution layer
The graph convolution layer outputs the potential representation of the nodes through convolution operations on the gene expression features and the adjacency matrix.
The formula for aggregation in the convolution layer is shown in Eq. ( 3 ) 35 :
where \(H_{k}\) denotes the node feature representation at the \(k-th\) layer of the node, the adjacency matrix after Laplace normalization is denoted as \({\tilde{D}}^{-\frac{1}{2}}{\tilde{A}}{\tilde{D}}^{-\frac{1}{2}}\) , \({\sigma }\) denotes the activation function \(\text {sigmoid}\) , W denotes the weight, and \(H_{k-1}\) denotes the node feature representation at the \(k-1th\) layer.
The nodes obtain the neighbor aggregation of the current order through the graph convolution operation and pass it to the next convolution layer. After the network training process, the node feature representation that reflects the features of the entire graph is ultimately obtained.
Transfer entropy
During the process of order-by-order aggregation, the neighbor information of nodes is continuously updated. However, this continuous updating leads to the loss of certain original neighbor information. As a result, it is challenging for the final node feature representation to fully capture the entire graph, leading to a decrease in the accuracy of the downstream task. To tackle this issue, measuring the degree of acceptance and retention of the current node feature representation following neighbor aggregation by Transfer Entropy, which measures the causal relationship between two time series by quantitatively describing the flow of information from one time series to another.
The value of Transfer Entropy indicates the strength of the causal relationship between the two time series, as shown in Eq. ( 4 ):
where X and Y denote two discrete time series, \(x_{n}\) and \(y_{n}\) are the discrete values of X and Y in the \(\text {n-th}\) instance, respectively, \(p(x_{n+1}|x_n,y_n)\) is the joint probability of \(x_{n}\) , \(x_{n+1}\) , \(y_{n}\) , and \(p(x_{n+1}\mid x_n)\) is the conditional probability of \(x_{n}\) and \(x_{n+1}\) .
In order to measure the information loss of node-by-node aggregation, the historical information of neighbor aggregation is retained during network training. Transfer Entropy is then calculated using both the historical information and the current value. The calculated value is saved and utilized for causal feature reconstruction calculations.
Causal feature reconstruction
After calculating the Transfer Entropy value, the neighbor aggregation with lower acceptance is weighted according to Eq. ( 5 ). Then, the neighbor information of each order is fused in the causal feature reconstruction module to obtain a more comprehensive and accurate representation of the causal node features. The larger the value of the Transfer Entropy, the less neighbor information is lost during transmission, and the lower weights assigned to that information. The structure of the causal feature reconstruction module is shown in Fig. 4 .

Structure of the causal reconstruction module.
where \(TE_{i\rightarrow j}^k\) denotes the Transfer Entropy of the accumulated i th-order neighbourhood information passed to the j th-order neighbourhood information at the k th training, and \(\zeta\) is a small constant, taken as \(\zeta =0.001\) to avoid computational errors when \(TE_{i\rightarrow j}^k\) could be zero.
The weighted neighborhood information is input into the MLP to complete the reconstruction. The Kullback-Leibler divergence is used to measure the discrepancy in the reconstruction, with the aim of minimizing the discrepancy throughout the neural network training process. Finally, the reconstruction feature \({\hat{Z}}\) is obtained, as shown in Eq. ( 6 ):
where \(concat(\cdot )\) denotes the collocation operation, \(MLP(\cdot ;\Omega )\) denotes the linear layer, \(\Omega\) is the parameter of the linear layer, \(Z_{j}\) is the j th-order neighbourhood aggregation, and \(Z_i^{\prime }\) is the feature weighted by Transfer Entropy.
The loss function of the entire CRGCN model consists of Binary Cross Entropy (BCEloss) and two Kullback-Leibler divergence, where the BCEloss function computes the classification error between labels build by known regulatory relationships and labels predicted by the model, both of the Kullback-Leibler divergence measure the reconstructed error and the weighted error of CRGCN, which could reinforce the causal information in neighbourhood aggregation. The function is shown as following:
where \({\hat{Z}}\) is the reconstruction causal feature, \(Z_j^{\prime }\) is the feature weighted by Transfer Entropy, \(Z_{j}\) is raw features exported from GCN, W denotes the weight and \(\lambda\) denotes the regularization factor.
After completing the causal feature reconstruction, the obtained reconstructed feature \(Z_j^{\prime }\) is normalized to smooth the training process and prevent gradient explosion. This is done in preparation for the subsequent link prediction task.
Link prediction module
After obtaining the causal reconstruction features of the nodes through the causal feature reconstruction module, the Preferential Preferential (PA) 36 is used to predict the similarity scores. These scores indicate the similarity between the current network node feature representations and the inferred network node feature representations. This method is computationally efficient, performs well in densely linked networks, and is particularly suitable for gene regulatory networks. The equation for the scores is ( 8 ):
where \({\hat{Z}}_i\) denotes current network node feature representation and \({\hat{Z}}_j\) denotes inferred network node feature representation.
Using link prediction, the inferred network is obtained by analyzing node-by-node networks and calculating the probability of connectivity between nodes. This process generates the neighborhood matrix of the predicted network, which represents the inferred GRN A .
Realisation implementation
The specific steps for predicting inferred GRN using GCN based on causal feature reconstruction are shown in Algorithm 1.

Inference of gene regulatory networks
Experimental process and result analysis
Data set and evaluation indicators.
The DREAM5 dataset provided by the DREAM CHALLENGES 37 and the mDC networks (Mouse dendritic cell) 38 are used in this paper. The specific information about the dataset is presented in Table 1 . The S.cerevisiae network has more genes, fewer samples and TFs, the true-positive edges are less than the true-negative edges, which induce class imbalances.
The implementation of gene regulatory network inference by GCN link prediction involved several steps. The hyperparameter for the Gaussian kernel function was set by several experiments. The Autoencoder hidden nodes were set to 805, 536 and 383 corresponding to the number of samples in the E.coli network, S.cerevisiae and mDC network, respectively. The Adam optimizer was used, with a learning rate of 0.001 which is chosen by experiments. Additional L2 regularization was applied during training to prevent parameter overfitting, the L2 rate is 0.001 which is chosen by experiments. After obtaining the features, they are fed into the GCN. The dataset was divided into a training set (70%) and a test set (30%). The Adam optimizer parameters is same as Gaussian-kernel Autoencoder.
In this paper, AUROC (Area Under the Receiver Operating Characteristic Curve) and AUPRC (Area Under the Precision–Recall Curve) are used as evaluation metrics for link prediction. AUROC represents the area under the curve with the axes of True Positive Rate (TPR) and False Positive Rate (FPR), while AUPRC represents the area under the curve with the axes of Precision and Recall.
where TP is the number of true positives, TN is the number of true negatives, FP is the number of false positives and FN is the number of true negatives.
In order to verify the effectiveness of the method, three experiments are set up to verify the effectiveness of the feature extraction method proposed in this paper, the effectiveness of causal feature reconstruction and the effectiveness of link prediction, separately.
Experiment 1: validating the effectiveness of feature extraction methods
To evaluate the effectiveness of the proposed feature extraction method in this paper, we chose a range of input features, including the original gene expression data, the sample expression features extracted solely by the Gaussian kernel function (GKF), the features obtained through singular value decomposition (SVD) 29 , the features obtained through non-negative matrix factorization (NMF) 28 , the fusion with the features extracted by the 1DCNN method 22 , and the features extracted by the Gaussian-kernel Autoencoder (gAE). For the link prediction task, a two-layer GCN was selected as the network model, and the number of iterations for network training was determined by E.coli , S.cerevisiae and mDC networks, and comparative results are depicted in Figs. 5 , 6 , 7 .

Comparison of results from different feature extraction methods in the E.coli network.

Comparison of results from different feature extraction methods in the S.cerevisiae network.

Comparison of results from different feature extraction methods in the mDC network.
Figures 5 , 6 , 7 demonstrate that the original gene expression features resulted in the lowest AUROC and AUPRC metrics for the E.coli network, the S.cerevisiae network and the mDC network. The NMF and SVD methods achieved higher metrics by compressing and filtering of the expression data. Using the GKF, the AUROC and AUPRC metrics improved to 0.804 and 0.801 in the E.coli network, 0.801 and 0.711 in the S.cerevisiae network, 0.656 and 0.642 in the mDC network, these metrics are higher than using the original data, 1DCNN, NMF, and SVD, indicating that the separable features can improve the accuracy of inferring gene regulatory networks.
In the E.coli network, using the gAE to extract features achieved the highest AUROC and AUPRC metrics, surpassing the GKF method by approximately 3% in the AUPRC. In the S.cerevisiae network, the AUROC metric for feature extraction by the gAE was 5.8% higher than using the GKF, however, the AUPRC metrics were slightly lower due to the imbalance in the categories of this network, which has more negative edges. In the mDC network, the AUROC metric and AUPRC metric surpassing the GKF method approximately by 13% and 19%, which achieved the highest AUROC and AUPRC metrics. Therefore, the gAE is able to mine deeper, more complex features to improve prediction accuracy compared to GKF. The result shows that the gAE feature extraction method is effective in the E.coli , the S.cerevisiae and the mDC network, which provides sufficient guidance for subsequent link prediction tasks.
In order to assess the reliability of the Gaussian-kernel Autoencoder in the separable features, and the effect of the Gaussian kernel parameter \(\sigma\) (in eqution(1)) in separable features and prediction results, the parameter \(\sigma\) was taken to be 0.1, 0.5, 1, 2, and 5, tested by a two-layer GCN and a GCN based on causal feature reconstruction (CRGCN), the results of the tests are shown in Table 2 .
From the Table 2 , it can be seen that when the parameter \(\sigma\) of the Gaussian kernel is taken as 1, the E.coli , the S.cerevisiae and the mDC network have the highest AUROC and AUPRC metrics.
The T-SNE method 39 is used to visually analyse the original features and the separable features. Figures 8 , 9 , 10 , 11 , 12 demonstrating that the deep and separable features are extracted by the gAE. In each Fig, the blue represents the E.coli network, the green represents S.cerevisiae network, the orange represents mDC network, and in each sub-graph of the Fig, the raw features are shown on the left and the separable features are shown on the right.

Visualization of the network features, when \(\sigma =0.1\) .

Visualization of the network features, when \(\sigma =0.5\) .

Visualization of the network features, when \(\sigma =1\) .

Visualization of the network features, when \(\sigma =2\) .
The parameter \(\sigma\) determines the distribution of the data in feature space, the larger \(\sigma\) , the features are leaded into more sparse space, made the features over-separated, conversely, the smaller \(\sigma\) , the features are leaded into more denser space, made the features unseparated. Both the larger \(\sigma\) and the smaller \(\sigma\) ineffectively extract separable features, resulting in lower AUROC and AUPRC metrics on CRGCN and GCN. The more properly separable features are extracted by Gaussian kernel when \(\sigma =1\) , therefore \(\sigma =1\) is selected in the subsequent experiments.

Visualization of the network features, when \(\sigma =5\) .
Overall, the method of extracting gene expression data into separable expression features is effective. Additionally, using the Autoencoders to combine these two features can better preserve the underlying information of the original expression data. This allows the graph neural network to obtain a more precise and comprehensive representation of node features during the node aggregation stage, ultimately enhancing the accuracy of the subsequent link prediction task.
Experiment 2: validating the effectiveness of causal feature reconstruction
In order to validate the effectiveness of the causal feature reconstruction method, the SVD, NMF, GKF, and gAE are selected as the methods for feature extraction. A two-layer GCN and a GCN based on causal feature reconstruction (CRGCN) are used as the network models for the link prediction task, and tested on E.coli and S.cerevisiae networks.

Comparison of results from different network models in the E.coli network.
The former four groups in Figs. 13 , 14 , 15 display the results of different feature extraction methods combined with GCN in the link prediction task, the latter four groups show the results of the methods combined with CRGCN.

Comparison of results from different network models in the S.cerevisiae network.

Comparison of results from different network models in the mDC network.
As shown in Figs. 13 , 14 , 15 , both the AUROC and AUPRC metrics showed significantly higher values in the latter four groups compared to the former four groups. In the E.coli network, compared to the gAE-GCN method, the gAE-CRGCN method improved the AUROC metrics by 9.5% and the AUPRC metrics with 3.2%. In S.cerevisiae network the AUROC metric improved with 7.4% and the AUPRC metric improved by 26%. Similarly, in mDC network the AUROC metric improved with 17.3% and the AUPRC metric improved by 15.4%.The results illustrate that using causal feature reconstruction can lead to a deeper causal features, which in turn improves the accuracy and precision of preferential connection prediction.
Overall, causal feature reconstruction enables the GCN model to obtain a more comprehensive representation of node features by enhancing the causal relationship between neighboring nodes at each order. It is able to capture deeper details from the gene expression features, ultimately improving the accuracy of link prediction, when combined with an effective feature extraction method for gene expression data.
Experiment 3: validating the effectiveness of link prediction using GCN based on Causal feature reconstruction
To make the model more accurate, the learning rate was chosen as 0.01, 0.005, 0.001, 1e−4 and 1e−5, the results are shown in Figs. 16 and 17 .

The AUROC with different learning rate.

The AUPRC with different learning rate.
From Figs. 16 and 17 , it can be seen that when the learning rate is chosen to 0.001, the model achieves the best AUROC and AUPRC, therefore, the learning rate is chosen to 0.001 in subsequent experiments.
To further validate the reliability and effectiveness of the gAE-CRGCN, 10-fold cross-validation is performed on the E.coli , the S.cerevisiae and the mDC networks, the results are shown in Figs. 18 , 19 , 20 .

10-fold cross-validation on the E.coli network.
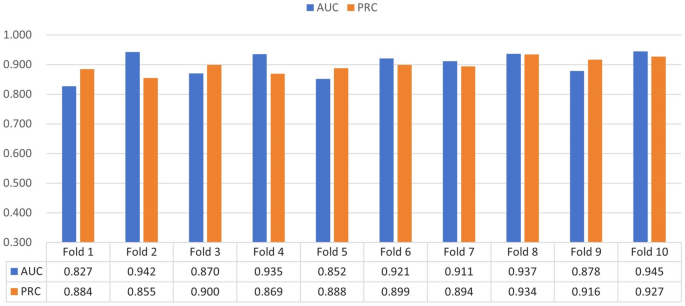
10-fold cross-validation on the S.cerevisiae network.

10-fold cross-validation on the mDC network.
In the 10-fold cross-validation experiment analysis, the E.coli , the S.cerevisiae , and the mDC networks are divided into 10 equal folds. The gAE-CRGCN is trained on 9 folds and tested on the remaining fold, the process is repeated 10 times, each time with a different fold to test, which helps to assess the performance and generalisation ability of the gAE-CRGCN. The Figs. 18 , 19 , 20 shown that the performers of the gAE-CRGCN model is stable within certain intervals.
Table 3 displays the AUROC and AUPRC scores for both existing methods and the methods proposed in this paper. As shown in Table 3 , it can be seen that the SVM method 40 demonstrates poor performance on large-scale biological networks and is unable to learn complex regulatory relationships. The RF method 41 achieves better results by constructing multiple decision tree models to infer biological networks. However, the AUPRC metric for the S.cerevisiae network is only 0.691, which is lower than that of the GNN method. This indicates that it is difficult for the RF algorithm to reliably infer the class-imbalanced networks. VGAE obtains new node feature representations by sampling the distribution of node feature representations using VGAE, however, data regeneration from the latent space has KL vanishing 36 , resulting in a poor metric. The GRGNN method combines the network skeleton predicted by known regulatory relationships, Pearson coefficients, and the network skeleton predicted by mutual information to obtain the input neighborhood matrix, GRDGNN 42 uses a multi-order neighborhood graph additionally. The GENELink 20 is composed of the Graph Attention Network, which incur huge computational and memory overhead than GCN, due to its graph-based attention mechanisms. The GNNLink 23 is a GCN-based interaction encoder, by capturing interdependencies between neighbors in the network to infer GRN.
The time consumption (second) of methods based on graph network is as shown in Table 4 .
From Table 4 , it can be seen that the proposed method has the second lowest running time, which means that the proposed method achieves better performance with less computational cost.
Network inference for the E.coli , the S.cerevisiae and the mDC was completed using a Gaussian-kernel Autoencoder with GCN based on causal feature reconstruction (gAE-CRGCN). The GRGNN and GRDGNN methods achieve higher AUROC metrics on E.coli and S.cerevisiae networks, by attaching extra network skeletons and obtaining input neighborhood matrices, however increasing the additional demand for data. The AUROC metric for the gAE-CRGCN method on the E.coli network was slightly lower than the GRGNN, however, the AUPRC metric was 6% higher than the GRGNN, 4.1% and 5.5% higher than GNNLink and GENELink, which are the state-of-the-art methods. Similarly, the AUROC metric of the gAE-CRGCN method on the S.cerevisiae network was slightly lower than GRDGNN, however, the AUPRC metric was 2.8% higher than GRDGNN, 6% and 4% higher than GNNLink and GENELink, the AUPRC is more valued in the GRN inference. The AUROC metric for the gAE-CRGCN method on the mDC network was 0.23% and 2% higher than GRGNN and GRDGNN, the AUPRC metrics was 8.6% and 3.5% higher than GNNLink and GENELink achieved the highest metrics. The gAE-CRGCN achieved the highest AUPRC in the three datasets, indicating that the proposed method has better prediction accuracy, due to the Causal Feature Reconstruction and Gaussian-kernel Autoencoder. The gAE-CRGCN method does not have any additional data requirements and improves the accuracy of node representations through causal reconstruction, which is capable of generating more accurate prediction results for class-imbalanced gene regulatory networks, with improved recall and precision.
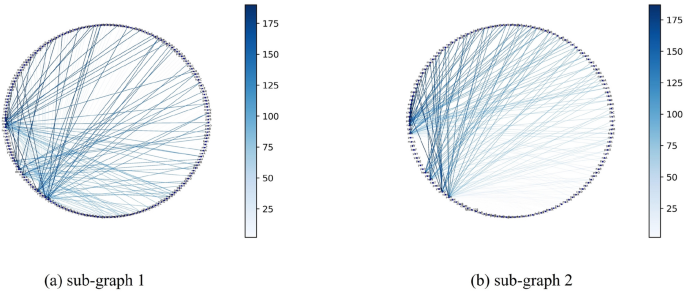
Sub-graph of the E.coli inferred network.

Sub-graph of the S.cerevisiae inferred network.
The sub-graph network are extracted from the inferred network and visualised as shown in are shown in Figs. 21 , 22 , 23 , which intended to show the details of the inferred network sub-graphs. It can be seen the different GRN have different densities of regulatory relationships. Figure 21 shows that a number of gene regulatory relationships in E.coli are dispersed among one another. As shown in Fig. 22 , a number of gene regulatory relationships in the E. coli network are dispersed among one another. As shown in Fig. 22 a, some genes like YLR121C, YJR141W, YNL156C, and YGR165W have rather more regulatory relationships, and as shown in Fig. 22 b gene YNL167C has the most regulatory relationships.
Overall, the gAE-CRGCN method has higher AUPRC scores, which implies the model has better precision and is more suitable for inferring the GRN. The gAE-CRGCN method enhances the node aggregation at each order, resulting in more detailed and comprehensive node feature representations. This is achieved by combining the fusion features extracted by a Gaussian-kernel Autoencoder. The enhanced node feature representations lead to higher similarity in predicting link priority connections, ultimately improving the accuracy of network inference. Experiments have confirmed that the method proposed in this paper is effective.

Sub-graph of the mDC inferred network.
GRN inference using graph GCN has become one of the advanced research. In order to improve the reliability and accuracy of GRN inference, the causal relationships of the node features gained by GCN should be tightened. In this paper, we propose a GCN for inferring GRN that is guided by causal information. The approach acquires the causal and comprehensive node representations to mitigate the loss of information during neighbor aggregation by reconstructing causal features. The Gaussian-kernel Autoencoder is proposed to extract significantly separable features from gene expression data, which improves the computational efficiency and reliability of causal feature reconstruction and the accuracy of inferring GRN. Experiments conducted on the DREAM5 dataset and the mDC dataset demonstrate that the approach proposed in this paper achieves superior prediction accuracy. Furthermore, it successfully reduce limitation of GCN in preserving the information of neighboring nodes at each order, resulting in improved accuracy. The proposed GCN link prediction method, which relies on causal feature reconstruction, enables the acquisition of node feature representations that possess causal feature. Consequently, this approach facilitates the construction of gene regulatory networks that are both reasonable and accurate, while also instilling a sense of credibility.
The proposed model can infer the gene regulatory network accurately, however, the model is impaired by class imbalance and known regulatory pairs insufficiency. In the future, we will fully exploit the priori knowledge of gene regulatory relationships to improve the performance with class imbalance, and explore integration of the multi-omics data to expand the biodata and information on regulatory relationships.
Data availability
The Dream5 dataset can be found at https://dreamchallenges.org/closed-challenges/. The mDC dataset can be found at https://zenodo.org/records/3701939.
Mochida, K., Koda, S., Inoue, K. & Nishii, R. Statistical and machine learning approaches to predict gene regulatory networks from transcriptome datasets. Front. Plant Sci. 9 , 1–7. https://doi.org/10.3389/fpls.2018.01770 (2018).
Article Google Scholar
Ahmed, S. S., Roy, S. & Kalita, J. Assessing the effectiveness of causality inference methods for gene regulatory networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 17 , 56–70. https://doi.org/10.1109/TCBB.2018.2853728 (2020).
Article PubMed Google Scholar
Ma, Q. et al. Uncovering mechanisms of transcriptional regulations by systematic mining of cis regulatory elements with gene expression profiles. BioData Min. 1 . https://doi.org/10.1186/1756-0381-1-4 (2008).
Park, J. et al. UPF1/SMG7-dependent microRNA-mediated gene regulation. Nat Commun . 10 . https://doi.org/10.1038/s41467-019-12123-7 (2019).
Ma, B., Fang, M. & Jiao, X. Inference of gene regulatory networks based on nonlinear ordinary differential equations. Bioinformatics . 36 , 4885–4893. https://doi.org/10.1093/bioinformatics/btaa032 (2020).
Article CAS PubMed Google Scholar
Friedman, N., Linial, M., Nachman, I. & Peer, D. Using Bayesian networks to analyze expression data. J. Comput. Biol. 7 (3–4), 601–620. https://doi.org/10.1089/106652700750050961 (2000).
Ajmal, H. B. & Madden, M. G. Dynamic Bayesian network learning to infer sparse models from time series gene expression data. IEEE/ACM Trans. Comput. Biol. Bioinform. 19 , 2794–2805. https://doi.org/10.1109/TCBB.2021.3092879 (2022).
Olsen, C., Meyer, P.E., & Bontempi, G. Inferring causal relationships using informationtheoretic measures. Proc. 5th Benelux Bioinf. Conf. (BBC09) (2009).
Haonan, F. NIMCE: a gene regulatory network inference approach based on multi time delays causal entropy. IEEE/ACM Trans. Comput. Biol. Bioinform. 19 , 1042–1049. https://doi.org/10.1109/TCBB.2020.3029846 (2020).
Sun, J., Taylor, D. & Bollt, E. M. Causal network inference byoptimal causation entropy. SIAM J. Appl. Dynamical Syst. 14 (1), 73–106 (2015).
Article MathSciNet Google Scholar
Sun, J. & Bollt, E. M. Causation entropy identifies indirect influences dominance of neighbors and anticipatory couplings. Phys. D: Nonlinear Phenom. 267 , 49–57. https://doi.org/10.1016/j.physd.2013.07.001 (2014).
Article ADS MathSciNet Google Scholar
Muzio, G. Biological network analysis with deep learning. Brief. Bioinform. 22 , 1515–1530. https://doi.org/10.1093/bib/bbaa257 (2021).
Li, W., Guo, Y., Wang, B. & Yang, B. Learning spatiotemporal embedding with gated convolutional recurrent networks for translation initiation site prediction. Pattern Recognit. 136 . https://doi.org/10.1016/j.patcog.2022.109234 (2023).
Liu, W. et al. MPCLCDA: predicting circRNA-disease associations by using automatically selected meta-path and contrastive learning. Brief. Bioinform. 24 (4), bba227. https://doi.org/10.1093/bib/bbad227 (2023).
Article CAS Google Scholar
Guo, Y., Zhou, D., Ruan, X. & Cao, J. Variational gated autoencoder-based feature extraction model for inferring disease-miRNA associations based on multiview features. Neural Netw. 165 , 491–505. https://doi.org/10.1016/j.neunet.2023.05.052 (2023).
Meroua, D. & Souham, M. Deep neural network for supervised inference of gene regulatory network. Model. Implement. Complex Syst. 64 , 149–157. https://doi.org/10.1007/978-3-030-05481-6_11 (2018).
Dan, M. L. A convolutional neural network for predicting transcription alregulators of genes in arabidopsis transcriptome data reveals classification based on positive regulatory interactions. bioRxiv. https://doi.org/10.1101/618926 (2019).
Scarselli, F., Gori, M. & Tsoi, A. C. The graph neural network model. IEEE Trans. Neural Netw. 20 , 61–80. https://doi.org/10.1109/TNN.2008.2005605 (2009).
Wang, J., Ma, A., Ma, Q., Dong, X. & Joshi, T. Inductive inference of gene regulatory network using supervised and semi-supervised graph neural networks. Comput. Struct. Biotechnol. J. 18 , 3335–3343. https://doi.org/10.1016/j.csbj.2020.10.022 (2020).
Article CAS PubMed PubMed Central Google Scholar
Chen, G. & Liu, Z. P. Graph attention network for link prediction of gene regulations from single-cell RNA-sequencing data. Bioinformatics 38 (19), 4522–4529. https://doi.org/10.1093/bioinformatics/btac559 (2022) ( PMID: 35961023 ).
Kipf, T.N. & Welling, M. Semi-Supervised Classification with Graph Convolutional Networks . https://doi.org/10.48550/arXiv.1609.02907 . (2016).
Ganeshamoorthy, S., Roden, L., Klepl, D. & He, F. Gene regulatory network inference through link Prediction using graph neural network. IEEE Signal Process. Med. Biol. Symp. (SPMB) . https://doi.org/10.1109/SPMB55497.2022.10014835 (2022).
Mao, G. et al. Predicting gene regulatory links from single-cell RNA-seq data using graph neural networks. Brief. Bioinform. 24 , bbad414. https://doi.org/10.1093/bib/bbad414 (2023).
Liu, Y., & Aviyente, S. The relationship between transfer entropy and directed information. 2012 IEEE Statistical Signal Processing Workshop (SSP). https://doi.org/10.1109/SSP.2012.6319809 . (2012).
Duan, Z., Xu, H., Huang, Y., Feng, J. & Wang, Y. Multivariate time series forecasting with transfer entropy graph. Tsinghua Sci. Technol. 28 , 141–149. https://doi.org/10.26599/TST.2021.9010081 (2023).
Zhang, J., Cao, J., Huang, W., Shi, X. & Zhou, X. Rutting prediction and analysis of influence factors based on multivariate transfer entropy and graph neural networks. Neural Netw. 157 , 26–38. https://doi.org/10.1016/j.neunet.2022.08.030 (2023).
An, J., Kim, K. and Kim, S. An algorithm for identifying differentially expressed genes in multiclass RNA-seq samples. 2014 International Conference on Big Data and Smart Computing (BIGCOMP). https://doi.org/10.1109/BIGCOMP.2014.6741402 . (2014).
Mirzal, A. NMF based gene selection algorithm for improving performance of the spectral cancer clustering. 2013 IEEE International Conference on Control System, Computing and Engineering . https://doi.org/10.1109/ICCSCE.2013.6719935 . (2013).
Fan, A., Wang, H., Xiang, H. & Zou, X. Inferring large-scale gene regulatory networks using a randomized algorithm based on singular value decomposition. IEEE/ACM Trans. Comput. Biol. Bioinform. 16 , 1997–2008. https://doi.org/10.1109/TCBB.2018.2825446 (2019).
Jayasumana, S., Hartley, R., Salzmann, M., Li, H. & Harandi, M. Kernel methods on Riemannian manifolds with Gaussian RBF Kernels. IEEE Trans. Pattern Anal. Mach. Intell. 37 , 2464–2477. https://doi.org/10.1109/TPAMI.2015.2414422 (2015).
Yang, Y., Tian, S., Yushan Qiu, P. & Zhao, Q. Z. MDICC: novel method for multi-omics data integration and cancer subtype identification. Brief. Bioinform. 23 . https://doi.org/10.1093/bib/bbac132 (2022).
Munquad, S. & Das, A. B. DeepAutoGlioma: a deep learning autoencoder-based multi-omics data integration and classification tools for glioma subtyping. BioData Min. 16 . https://doi.org/10.1186/s13040-023-00349-7 (2023).
Wang, C.-C., Li, T.-H., Huang, L. & Chen, X. Prediction of potential miRNA-disease associations based on stacked autoencoder. Brief. Bioinform. 23 . https://doi.org/10.1093/bib/bbac021 (2022).
Li, X. et al. MoGCN: A multi-omics integration method based on graph convolutional network for cancer subtype analysis. Front. Genet. 13 . https://doi.org/10.3389/fgene.2022.806842 (2022).
Kipf, T. N., Welling, M. Semi-supervised Classification With Graph Convolutional Networks. ICLR. https://openreview.net/forum?id=SJU4ayYgl . (2017).
Kumar, A., Singh, S. S., Singh, K. & Biswas, B. Link prediction techniques, applications, and performance: A survey. Phys. A . 533 . https://doi.org/10.1016/j.physa.2020.124289 (2020).
Marbach, D. et al. Wisdom of crowds for robust gene network inference. Nat. Methods 9 (8), 796–804 (2012).
Pratapa, A. et al. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods . 17 , 147–154. https://doi.org/10.1038/s41592-019-0690-6 (2020).
Chourasia, P., Ali, S., & Patterson, M. Informative Initialization and Kernel Selection Improves t-SNE for Biological Sequences. 2022 IEEE International Conference on Big Data (Big Data) . https://doi.org/10.1109/BigData55660.2022.10020217 . (2022).
Lazzarini, N. et al. Functional networks inference from rule-based machine learning models. BioData Min. 9 . https://doi.org/10.1186/s13040-016-0106-4 (2016).
Li, J. et al. Detecting gene-gene interactions using a permutation-based random forest method. BioData Min. 9 . https://doi.org/10.1186/s13040-016-0093-5 (2016).
Liao, Q., Wu, X., Xie, X., Wu, J., Qiu, L., & Sun, L. Adversarial residual variational graph autoencoder with batch normalization. 2021 IEEE Sixth International Conference on Data Science in Cyberspace (DSC). https://doi.org/10.1109/DSC53577.2021.00013 . (2021).
Zhenyu, G. and Wanhong, Z. An efficient inference schema for gene regulatory networks using directed graph neural networks. Proceedings of the 42nd Chinese Control Conference . https://doi.org/10.23919/CCC58697.2023.10240472 . (2023)
Download references
This work was supported in part by the National Natural Science Foundation of China under Grant 61702410.
Author information
Authors and affiliations.
School of Automation and Information Engineering, Xi ’an University of Technology, No.5, Jinhua South Road, Xi’an, 710048, Shaanxi, China
Ruirui Ji, Yi Geng & Xin Quan
Key Laboratory of Shaanxi Province for Complex System Control and Intelligent Information Processing, Xi’an, 710048, Shaanxi, China
You can also search for this author in PubMed Google Scholar
Contributions
R.J. conceived methodology and modified the manuscript. Y.G. wrote the manuscript and implemented the software. X.Q. performed the validation and visualization. All authors have read and approved the manuscript.
Corresponding author
Correspondence to Ruirui Ji .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Ethical approval and consent to participate
This study did not include the use of any animals, human or otherwise, so did not require ethical approval. Informed consent was obtained from all individuals included in this study.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Ji, R., Geng, Y. & Quan, X. Inferring gene regulatory networks with graph convolutional network based on causal feature reconstruction. Sci Rep 14 , 21342 (2024). https://doi.org/10.1038/s41598-024-71864-8
Download citation
Received : 13 June 2024
Accepted : 02 September 2024
Published : 12 September 2024
DOI : https://doi.org/10.1038/s41598-024-71864-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Gene regulatory network
- Causal relationship
- Graph convolutional network
- Link prediction
- Autoencoder
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
This browser is no longer supported.
Upgrade to Microsoft Edge to take advantage of the latest features, security updates, and technical support.
/experimental:module (Enable module support)
- 2 contributors
Enables experimental compiler support for C++ Standard modules. This option is obsolete for C++20 standard modules in Visual Studio version 16.11 and later. It's still required (along with /std:c++latest ) for the experimental Standard library modules.
/experimental:module [ - ]
In versions of Visual Studio before Visual Studio 2019 version 16.11, you can enable experimental modules support by use of the /experimental:module compiler option along with the /std:c++latest option. In Visual Studio 2019 version 16.11, module support is enabled automatically by either /std:c++20 or /std:c++latest . Use /experimental:module- to disable module support explicitly.
This option is available starting in Visual Studio 2015 Update 1. As of Visual Studio 2019 version 16.2, C++20 Standard modules aren't fully implemented in the Microsoft C++ compiler. Modules support is feature complete in Visual Studio 2019 version 16.10. You can use the modules feature import the Standard Library modules provided by Microsoft. A module and the code that consumes it must be compiled with the same compiler options.
For more information on modules and how to use and create them, see Overview of modules in C++ .
To set this compiler option in the Visual Studio development environment
Open the project's Property Pages dialog box. For details, see Set C++ compiler and build properties in Visual Studio .
Set the Configuration drop-down to All Configurations .
Select the Configuration Properties > C/C++ > Language property page.
Modify the Enable C++ Modules (experimental) property, and then choose OK .
/headerUnit (Use header unit IFC) /exportHeader (Create header units) /reference (Use named module IFC) /translateInclude (Translate include directives into import directives) /Zc (Conformance)
Was this page helpful?
Additional resources
Navigation Menu
Search code, repositories, users, issues, pull requests..., provide feedback.
We read every piece of feedback, and take your input very seriously.
Saved searches
Use saved searches to filter your results more quickly.
To see all available qualifiers, see our documentation .
- Notifications You must be signed in to change notification settings
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement . We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
ExperimentalWarning: Importing JSON modules #51347
webdevnerdstuff commented Jan 3, 2024
| 20.9.0 & 20.10.0 Darwin foobar.local 22.6.0 Darwin Kernel Version 22.6.0: Thu Nov 2 07:43:57 PDT 2023; root:xnu-8796.141.3.701.17~6/RELEASE_ARM64_T6000 arm64 (same result with ) Every time you run a package script it's throwing this warning in the console. It should not be throwing a warning if there is nothing that is being used in that warning. As it seems the trace shows the warnings coming from . There are places in the code where I am importing a json file, but I also tested by removing all of those imports, and the output of the warning is the same. So I'm not sure where and/or why it's throwing this warning. Is this a false positive? |
| The text was updated successfully, but these errors were encountered: |
aduh95 commented Jan 3, 2024
| Well unless you're able to reproduce without using any external dependency, I'm going to assume one of your deps is importing a JSON module, which is still experimental hence the runtime warning you're seeing. You can add to your env to get more information on what is being imported at the time the warning gets emitted. |
Sorry, something went wrong.
| There are a few problems with how the warning is being handled currently in my opinion that could use improvements. I now realize it is the warning itself that is the issue here. Not using any external dependencies is not a helpful response as that is not a realistic or real world situation, nor does it help solve the issue, as removing packages would just cause an avalanche of other errors. Your first sentence also comes across as a bit condescending, but I'm just going to assume it's a language barrier situation and move on. I think it would be more useful if the warning mentioned that it could be an external dependency causing the issue. I spent a good amount of time combing through my own code looking in the wrong direction trying to figure out what may have caused the warning. If in a larger team, it could cause multiple people looking in the wrong direction and wasting a lot of time as well. It think it would also be helpful if the warning included your suggestion to use , which is a much better suggestion to figure out the problem. The message given includes: When you run that command, it makes it seem like it's a node problem, not any external dependency. Perhaps I read the warning wrong at first, but it's message implies it will help find what's causing the warning in my code. Another thing it could imply is that it will just tell you where in the node source code that is throwing that warning, which is completely useless when problem solving what's causing the warning in the project. I want to know what's causing the issue so I can fix it and/or perhaps find another solution in the dependency causing it, not what code is throwing the warning within node as that is useless information, unless I'm debugging the actual warning message itself. I would ask to the node team to make some adjustments to the warning to help avoid a lot of wasted time spent trying to find an issue while looking in the wrong direction. It could save time spent debugging and potentially an entire team doing the same, searching the internet for a solution, submitting an issue to GitHub, reading the response, spending time responding, and would have saved any Contributor to node, time reading/responding to the submitted issue. |
- 👍 1 reaction
johnleider commented Jan 3, 2024
| I can accept that it's experimental, but is there a way to suppress the warning? |
- 👍 3 reactions
jasnell commented Jan 3, 2024
| With older versions, will turn off all warnings. Newer versions also support where is the specific code associated with an individual warning (if there is one) |
- 🎉 1 reaction
webdevnerdstuff commented Jan 3, 2024 • edited Loading
| It looks like that was added in . That would be nice to have in the LTS version. For now I'll most likely just add (or just depending) into the script. |
| Apologies, that was not my intention. I was meant as an answer for “is this a false positive?” (or just depending) into the script.Note that disabling warnings means you’re more likely to miss important information, you might want to consider avoiding use of experimental API instead (or accept to see the warning). If you are writing something that is going to be run by someone else, have in mind that removing the warning might be a disservice to your users, as experimental APIs are not bound to semver rules and could break anytime they update their version of Node.js. |
- 👍 4 reactions
| I had a feeling it was just a language difference in how the sentence came across. No worries. In my situation where this came up, it is a component for another library. So it's unlikely this will affect them when it comes to working on this component, which I don't anticipate anybody wanting to help with this one. The package that's causing the warning is not something I can do without (at least not without difficulty), and/or would be used by the users in their own project as well, so as long as they don't silent the warning, they will still get it. Which brings me back to the message could use a little bit of tweaking in it's wording to help avoid confusion for other users when this comes up. |
aduh95 commented Jan 4, 2024
| Not sure how I feel about promoting usage in such warning, it's meant as a tool for Node.js core devs, not for end-users. We could try to add the information to the warning of which import triggered the warning, PRs welcome. In the mean time, I think you should report to the dependency you are using that they should document they use experimental syntax, and that users should expect to see that warning, and what that means in terms of support from Node.js updates (and how to silence the warning if that's what you want). |
webdevnerdstuff commented Jan 4, 2024
| Adding which import triggered the warning sounds like an excellent idea as that would get straight to the source of the problem. I wouldn't know where to begin to even attempt to submit a PR for it. If/when I get some free time, I'll definitely look into it and give it a try if someone else doesn't get to it first. I somewhat agree about promoting , as that could confuse people who are not as technical and overwhelm them with the output it gives not knowing what they should be looking for. At the very least, I think adding a message saying it could be an external dependency would be very helpful so people can possibly avoid having to dig through potentially hundreds of files looking for something that might not exist in their own code. |
liudonghua123 commented Jan 16, 2024
| I add a shebang like this to make the bin script working without showing the warnings. And it works for windows too. |
- 👍 8 reactions
shellscape commented May 11, 2024
| thanks for that workaround The clusterfck that is ESM in Node never ceases to amaze. |
petersem commented May 14, 2024 • edited Loading
| I did the following to work around the warning. |
- 👍 17 reactions
No branches or pull requests
- Stack Overflow for Teams Where developers & technologists share private knowledge with coworkers
- Advertising & Talent Reach devs & technologists worldwide about your product, service or employer brand
- OverflowAI GenAI features for Teams
- OverflowAPI Train & fine-tune LLMs
- Labs The future of collective knowledge sharing
- About the company Visit the blog
Collectives™ on Stack Overflow
Find centralized, trusted content and collaborate around the technologies you use most.
Q&A for work
Connect and share knowledge within a single location that is structured and easy to search.
Get early access and see previews of new features.
Experimental decorators warning in TypeScript compilation
I receive the warning...
Experimental support for decorators is a feature that is subject to change in a future release. Set the 'experimentalDecorators' option `to remove this warning.
... even though my compilerOptions in tsconfig.json have the following settings:
What is weird is that some random classes that use decorators do not show that warning but the rest in the same project do.
What could cause such behavior in the TypeScript compiler?
- visual-studio-code
- 85 Have you tried restarting VS Code? I've found that's necessary after making tsconfig.json changes sometimes. – David Sherret Commented Jul 8, 2016 at 16:26
- 17 By chance if someone else runs into this that is using VS Professional, not VS Code, you may have added a .ts file to an angular project manually; if so, the default TS compilation is conflicting with Angular CLI. Right-click the file -> Properties -> Build Action : None. Then restart VS if needed. – pbristow Commented Jul 19, 2018 at 14:16
- 9 VS Code restart helped me solve the issue. – CMA Commented Nov 6, 2018 at 6:28
- 4 As @paulsm4 said, the problem showed up in my case after starting VSCode in the wrong directory. You want to start VSCode in the directory where tsconfig.json is for your project. – ebhh2001 Commented Sep 23, 2019 at 18:58
- 3 Closing project and re-open the project solved my problem. – Om Prakash Gupta Commented Oct 21, 2020 at 11:59
40 Answers 40
I've to add the following in the settings.json file of vscode to remove the warning.
VSCode -> Preferences -> Settings

As Clepsyd pointed out, this setting had been deprecated. You need to use now

- 5 ctrl + , shortcut key to open usersettings. on usersettings click 3 dots ( ... ) then on pop up click Open settings.json to open settings.json – suhailvs Commented Oct 3, 2018 at 9:37
- I needed to do this. The tsconfig.json solution worked fine until I added a new service, but this time deeper as usual in the directory tree. (app->component->xyz_comp->sub_component->sub_service) – al-bex Commented Jul 13, 2020 at 11:59
- 33 This settings has been deprecated. You should now use: "js/ts.implicitProjectConfig.experimentalDecorators": true – Clepsyd Commented Dec 15, 2020 at 15:45
- "js/ts.implicitProjectConfig.experimentalDecorators": true .. fixed my issue. Looks like the other approach is depricated. – Ahadu Melesse Commented Jun 29, 2022 at 4:01
Although VS Code is a great editor for TypeScript projects, it needs a kick every now and again. Often, without warning, certain files cause it to freak out and complain. Mostly the fix seems to be to save and close all open files, then open tsconfig.json . After that you should be able to re-open the offending file without error. If it doesn't work, lather, rinse, and repeat.
If your tsconfig.json specifies its source files using the files array, IntelliSense will only function correctly if the file in question is referenced such that VS Code can find it by traversing the input file tree.
Edit: The 'reload window' command (added ages ago now) should solve this problem once and for all.
- 6 The answer was not really relevant to my case but somehow helped my figure out my problem. I am using VS 2015 and between changing the xproj and tsconfig file, I missed the fact that the folder with my script files was listed in the exclude section of tsconfig . Hope this helps someone. – Stefan Balan Commented Sep 3, 2016 at 18:10
- 36 File > Close folder > Open folder worked for me. This happened after I restarted the machine without properly closing vscode. – Adrian Moisa Commented Mar 24, 2017 at 12:28
- 3 What if i am not using typescript? Only javascript es6? – Playdome.io Commented Apr 22, 2017 at 22:17
- 10 I'm not sure when it was added but the command palette now has "Reload Window", which is ideal for this sort of thing. – Coderer Commented Aug 7, 2017 at 6:46
- 7 1. Go to File -> Preferences -> Settings. 2. Search "experimentalDecorators" 3. Check Enable/disable experimentalDecorators 4. Press Ctrl + S to save settings. – Syed Nasir Abbas Commented Jul 14, 2020 at 18:54

- 1 For me, the issue appeared on version 1.37 and your solution solved it. – Stephane Commented Aug 10, 2019 at 20:08
Please follow the below step to remove this warning message.

Step 1: Go to setting in your IDE then find or search the experimentalDecorators .

Step 2: then click on checkbox and warning has been remove in your page.

This error also occurs when you choose "src" folder for your workspace folder.
When the root folder, folder where the "src", "node_modules" are located is chosen, the error disappears
- 1 In case the above was not comprehensible: Vscode must load the folder where the config file is to know about it. If you want to load a folde further down I guess you could write a new config file that would be valid from that folder and below. – LosManos Commented Mar 16, 2020 at 13:14
inside your project create file tsconfig.json , then add this lines
In VSCode, Go to File => Preferences => Settings (or Control+comma) and it will open the User Settings file. Search "javascript.implicitProjectConfig.experimentalDecorators": true and then check the checkbox for experimentalDecorators to the file and it should fix it. It did for me.

have to add typescript.tsdk to my .vscode/settings.json :
- 5 This solved the issue for me, but that line goes into .vscode/settings.json as per this – Fran Rodriguez Commented Sep 20, 2016 at 16:42
- 1 Also solved it for me, but I didn't have the .vscode folder (I don't know why - I'm a backend dev, leave me alone!), so I created one in the root folder with said settings.json file. – Aage Commented May 10, 2017 at 6:13
- Thank you!! How on earth did you figure this out? – raphisama Commented Jul 17, 2020 at 3:48
- Ahem! Ahem! Sometimes, the path to node_modules could not be resolved. If that is the case we need to give the full relative path to the same like "typescript.tsdk": "./node_modules/typescript/lib" – sjsam Commented Oct 3, 2021 at 6:19
I get this warning displayed in vscode when creating a new Angular service with the
syntax (rather than providing the service in app.module.ts).
The warning persists until I reference the new service somewhere in the project. Once the service gets used the warning goes away. No typescript configuration or vscode settings changes necessary.
- I my case it was solved also when I reference the new service somewhere in the proyect, thanks! – Dylan Moguilevsky Commented Jun 10, 2022 at 13:10
For me, this error "Experimental support for decorators is a feature that is subject to change in a future release. (etc)" only happened in VS Code in an Angular project and only when creating a new Service.
The solution above: "In Visual Code Studio Go to File >> Preferences >> Settings, Search "decorator" in search field and Checking the option JavaScript › Implicit Project Config: Experimental Decorators" solved the problem.
Also, stopping the ng serve in the terminal window and restarting it made the error disappear after recompile.
This answer is intended for people who are using a Javascript project and not a Typescript one. Instead of a tsconfig.json file you may use a jsconfig.json file.
In the particular case of having the decorators warning you wan write inside the file:
Fort the buggy behaviors asked, it's always better to specify the "include" in the config file, and restart the editor. E.g.
- 1 Just in case, in either language it's important that the files you are working with are inclided in the "include", for example "app/*/*.ts" or "app/*/*.js", etc. That solved my problem. – reojased Commented Jul 19, 2020 at 7:15
Open settings.json file in the following location <project_folder>/.vscode/settings.json
or you can open the file from the menu as mentioned below
VSCode -> File -> Preferences -> Workspace Settings

Then add the following lines in settings.json file
That's all. You will see no warning/error regarding ' experimentalDecorators '
- 12 I got "Unknown configuration setting" after adding "enable_typescript_language_service" using vscode 1.8.1 – chubao Commented Jan 22, 2017 at 12:35
- 11 This is HORRIBLE advice if your project uses Typescript! – paulsm4 Commented Jan 3, 2019 at 17:22
- 2 Disabling "enable_typescript_language_service" would effectively turn off any live linting TypeScript offers please avoid this suggestion. – Jessy Commented Nov 14, 2019 at 15:23
- This is worked for me. for more info refer below link also. ihatetomatoes.net/… – Kodali444 Commented Dec 25, 2020 at 12:02
Will solve this problem.
- 1 @nadya: settings.json (either user or workspace). You can also use the gui settings, as this answer does – Frank N Commented Dec 12, 2019 at 12:00
Add following lines in tsconfig.json and restart VS Code.
STEP 1: Press ctrl + , in VS code
STEP 2: Enter 'js/ts.implicitProjectConfig: Experimental Decorators' in search box
STEP 3: check the checkbox related to the search

If you are using cli to compile *.ts files, you can set experimentalDecorators using the following command:
I had this problem recently under Visual Studio 2017 - turned out it was caused by a "feature" of VS - ignoring tsconfig.json when Build action is not set to Content .
So changing the Build action to Content and reloading the solution solved the problem.
Not to belabor the point but be sure to add the following to
- Workspace Settings not User Settings
under File >> Preferences >> Settings
"javascript.implicitProjectConfig.experimentalDecorators": true
this fixed the issue for me, and i tried quite a few suggestions i found here and other places.
- Open VScode.
- Press ctrl+comma
- Search about experimentalDecorators
I had this error with following statement
Experimental support for decorators is a feature that is subject to change in a future release. Set the 'experimentalDecorators' option in your tsconfig or jsconfig to remove this warning.ts(1219)
It was there because my Component was not registered in AppModule or (app.module.ts) i simply gave the namespace like
import { abcComponent } from '../app/abc/abc.component';
and also registered it in declarations
- I get the exact same error after creating a new file with "@Injectable() export class MyService {}" in it. Nothing to add to AppModule (ng10) – Wolf359 Commented Jul 18, 2020 at 18:23
- i had the same issue – Thomas Martin Commented Aug 27, 2020 at 16:52
For the sake of clarity and stupidity.
1) Open .vscode/settings.json.
2) Add "typescript.tsdk": "node_modules/typescript/lib" on it.
3) Save it.
4) Restart Visual Studio Code.
If you are working in Visual studio. You can try this fix
- Unload your project from visual studio
- Go to your project home directory and Open "csproj" file.
Add TypeScriptExperimentalDecorators to this section as shown in image

- Reload the project in Visual studio.
see more details at this location.
- Did you really use a screen shot of the text? – Mason240 Commented Jun 23, 2021 at 15:48
- yes, its screen shot from working project – kumar chandraketu Commented Jun 23, 2021 at 16:03
- Although I kindly want to downvote because of the screenshot, it gave me the solution as I don't use VS Code but VS 2022 and it was driving me nuts that tsconfig was fine and VS was still complaining about it. This one is nonsense from ms: " If you have a project file, tsconfig will not be honored. the project file takes precedence. " link – Perrier Commented Feb 22, 2022 at 10:14
Please check you oppened in your VS Code the folder of the entire project and not only the src folder, because if you open only the src, then ts.config.json (located in the project folder) file will not be in scope, and VS will not recognize the experimental decorators parameters.
In my case this fixed all the problems related to this issue.
- This was also my issue. Can you see node_modules? Can you see tsconfig.json? If not, open a new vsCode window, close the old window, choose file-->open folder, and make sure you select the parent folder of src and node_modules , rather than selecting just src . – Kyle Vassella Commented Feb 13, 2019 at 23:40
in my case I solved this issue by setting "include": [ "src/**/*"] in my tsconfig.json file and restarting vscode. I've got this solution from a github issue: https://github.com/microsoft/TypeScript/issues/9335
I used React and Nest for my project. Had this error displayed in the backend, but adding those two lines to react's tsconfig.json fixed the issue for some reason. Furthermore, everything above did not work for me
Open entire project's folder instead of project-name/src
tsconfig.json is out of src folder
I faced the same issue while creating an Injectable Services in Angular 2. I have all the things at place in tsconfig.json .Still I was getting this error at ColorsImmutable line.
And fix was to register the Service at module Level or Component Level using the providers array. providers:[ColorsImmutable ],
If you using Deno JavaScript and TypeScript runtime and you enable experimentalDecorators:true in tsconfig.json or the VSCode ide settings. It will not work. According to Deno requirement, you need to provide tsconfig as a flag when running a Deno file. See Custom TypeScript Compiler Options
In my particular case I was running a Deno test and used.
If it is a file, you have something like
my tsconfig.json
- I'm already doing this and yet no luck, as they state in the docs experimental features will not work, – Ezzabuzaid Commented May 25, 2020 at 16:54
- It will work. See updated answer of exactly the content of my tsconfig.json. Then see this link github.com/manyuanrong/dso/issues/18 – tksilicon Commented May 25, 2020 at 17:18
I resolved the same issue in the VS Code Version:1.76.0 and fixed it by following these steps.

- Then click on Configure 'TypeScript' language based settings...

- add line number 3 if it is not there. and save it:
Not the answer you're looking for? Browse other questions tagged typescript decorator visual-studio-code or ask your own question .
- The Overflow Blog
- One of the best ways to get value for AI coding tools: generating tests
- The world’s largest open-source business has plans for enhancing LLMs
- Featured on Meta
- User activation: Learnings and opportunities
- Site maintenance - Mon, Sept 16 2024, 21:00 UTC to Tue, Sept 17 2024, 2:00...
- What does a new user need in a homepage experience on Stack Overflow?
- Announcing the new Staging Ground Reviewer Stats Widget
Hot Network Questions
- Remove all punctuation AND the values after it at end of string in R
- Should I change advisors because mine doesn't object to publishing at MDPI?
- How to best characterize the doctrine deriving from Palsgraf?
- ASCII 2D landscape
- How to deal with coauthors who just do a lot of unnecessary work and exploration to be seen as hard-working and grab authorship?
- If Act A repeals another Act B, and Act A is repealed, what happens to the Act B?
- Negating a multiply quantified statement
- "Famous award" - "on ships"
- O(nloglogn) Sorting Algorithm?
- zsh completion - ignore executable files with no dot in the name
- Is it defamatory to publish nonsense under somebody else's name?
- Fast leap year check
- How to decrease by 1 integers in an expl3's clist?
- Why was Esther included in the canon?
- VBA: Efficiently Organise Data with Missing Values to Achieve Minimum Number of Tables
- Twists of elliptic curves
- Is Produce Flame a spell that the caster casts upon themself?
- Why is resonance such a widespread phenomenon?
- Definition of annuity
- How can I analyze the anatomy of a humanoid species to create sounds for their language?
- Drill perpendicular hole through thick lumber using handheld drill
- How can I send instance attributes from Geometry Nodes to Shading Editor?
- Is it true that before European modernity, there were no "nations"?
- Why does Sfas Emes start his commentary on Parshat Noach by saying he doesn't know it? Is the translation faulty?

COMMENTS
Error: Module uses experimental features on versions.tf line 3, in terraform: 3: experiments = [module_variable_optional_attrs] Experimental features are intended only for gathering early feedback on new language designs, and so are available only in alpha releases of Terraform.
Proposal. 1.3 concludes the module_variable_optional_attrs experiment and errors when you use the old experiment flag, plus it removes the defaults() function that was available as part of that experiment.. I propose that 1.3 be a bridge release where it emits a deprecation warning about the experiment concluding, and that it retain the defaults() function ONLY when the module_variable ...
In this article. The Experimental Features support in PowerShell provides a mechanism for experimental features to coexist with existing stable features in PowerShell or PowerShell modules. An experimental feature is one where the design isn't finalized. The feature is available for users to test and provide feedback.
You have to declare in your module that you're using the experiment: terraform {. # Optional attributes and the defaults function are. # both experimental, so we must opt in to the experiment. experiments = [module_variable_optional_attrs] } And then you would use it in your case like this: variable "list_of_users" {.
It is interesting that in this particular case there is some overlap between the experimental design and the final design, but experimental features are not part of the language and any module using them should expect to become "broken" either by future iterations of the experiment or by the experiment concluding and that experimental ...
╷ │ Warning: Experimental feature "module_variable_optional_attrs" is active │ │ on versions.tf line 3, in terraform: │ 3: experiments = [module_variable_optional_attrs] │ │ Experimental features are subject to breaking changes in future minor or │ patch releases, based on feedback. │ │ If you have feedback on the design of ...
We do not recommend using experimental features in Terraform modules intended for production use. In order to make that explicit and to avoid module callers inadvertently depending on an experimental feature, any module with experiments enabled will generate a warning on every terraform plan or terraform apply. If you want to try experimental ...
The Experimental Attribute. Use the Experimental attribute to declare some code as experimental.. Use the following syntax to declare the Experimental attribute providing the name of the experimental feature and the action to take if the experimental feature is enabled: [Experimental(NameOfExperimentalFeature, ExperimentAction)] For modules, the NameOfExperimentalFeature must follow the form ...
The Optional Object type attribute, was in beta for quite some time since Terraform 0.14 and now in Terraform 1.3 (released end of September 2022) it's GA. So to get this straight, this is not a new feature, but now it is 100% ready to be used in production use cases. When you are building a generic module and you want to offer a lot of ...
The Get-ExperimentalFeature cmdlet returns all experimental features discovered by PowerShell. Experimental features can come from modules or the PowerShell engine. Experimental features allow users to safely test new features and provide feedback (typically via GitHub) before the design is considered complete and any changes can become a breaking change.
But even that would require every module author to change their code ^^^ This doesn't make any sense from the perspective that if a development team depends on dozens of modules from various vendors that happen to be using the experimental module_variable_optional_attrs feature that they need to wait for every single module to update their code to 1.3.x in order to be usable.
Warning: Experimental feature "variable_validation" is active on prod.tf line 28, in terraform: 28: experiments = [variable_validation] Experimental features are subject to breaking changes in future minor or patch releases, based on feedback. If you have feedback on the design of this feature, please open a GitHub issue to discuss it.
Use type="module" in package.json, experimental modules and specify extensions with specifier-resolution like this: node --experimental-modules --es-module-specifier-resolution=node server.js. Don't use specifier-resolution, you'll have to specify the extension of your files every where. Update (from comment), for Node v18:
The Enable-ExperimentalFeature cmdlet enables experimental features by adding the named experimental features to the powershell.config.json settings file read on PowerShell startup. This cmdlet was introduced in PowerShell 6.2. Note. Any changes to experimental feature state only takes effect on restart of PowerShell.
In this example, utils.js shares the greet function, and my_script.js uses it. The <script type="module"> tag makes sure the browser knows my_script.js is a module.. Important things to know: Script Order: When you use multiple <script type="module"> tags, the browser runs them in the order they appear in the HTML. This ensures that everything loads in the right order.
You signed in with another tab or window. Reload to refresh your session. You signed out in another tab or window. Reload to refresh your session. You switched accounts on another tab or window.
After obtaining the causal reconstruction features of the nodes through the causal feature reconstruction module, the Preferential Preferential (PA) 36 is used to predict the similarity scores ...
In versions of Visual Studio before Visual Studio 2019 version 16.11, you can enable experimental modules support by use of the /experimental:module compiler option along with the /std:c++latest option. In Visual Studio 2019 version 16.11, module support is enabled automatically by either /std:c++20 or /std:c++latest.
ExperimentalWarning: Importing JSON modules is an experimental feature and might change at any time ryoppippi/unplugin-typia#212. Closed. Caesarovich mentioned this issue on Jul 16. 8.0 will expose ExperimentalWarning while use node 20+ sindresorhus/boxen#98. Closed. holic mentioned this issue 2 hours ago.
ERROR: Running multiple emulators with the same AVD is an experimental feature for ubuntu in android Studio? Hot Network Questions Is it feasible to create an online platform to effectively teach college-level math (abstract algebra, real analysis, etc.)?
Experimental support for decorators is a feature that is subject to change in a future release. Set the 'experimentalDecorators' option in your tsconfig or jsconfig to remove this warning.ts(1219) It was there because my Component was not registered in AppModule or (app.module.ts) i simply gave the namespace like